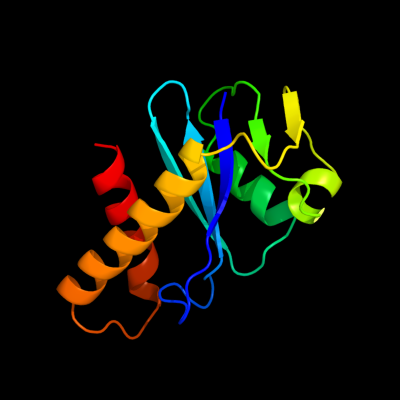
1 d1cxqa_
98.0
15
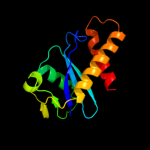
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain2 d1hyva_
98.0
14
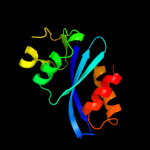
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain3 d1exqa_
97.6
11
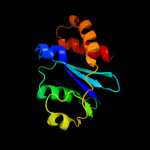
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain4 d1asua_
97.4
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain5 c1c0mA_
97.3
18
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
6 c1ex4A_
97.1
14
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
7 c3nf9A_
96.9
17
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
8 d1c0ma2
96.9
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain9 c3kksB_
96.8
21
PDB header: dna binding proteinChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
10 c3f9kV_
96.6
13
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
11 c1k6yB_
96.5
16
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
12 d1c6va_
96.4
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain13 c3dlrA_
93.1
11
PDB header: transferaseChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain from pfv2 integrase
14 c3hpgC_
92.7
14
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
15 c3l2tB_
83.4
9
PDB header: recombination/dnaChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
16 d1bcoa2
49.1
4
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain17 c1bcoA_
27.8
4
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
18 c1xb4C_
27.0
38
PDB header: unknown functionChain: C: PDB Molecule: hypothetical 23.6 kda protein in yuh1-ura8PDBTitle: crystal structure of subunit vps25 of the endosomal2 trafficking complex escrt-ii
19 d1xb4a1
24.1
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain20 c3he5D_
23.2
30
PDB header: de novo proteinChain: D: PDB Molecule: synzip2;PDBTitle: heterospecific coiled-coil pair synzip2:synzip1
21 c3cuqD_
not modelled
21.1
47
PDB header: protein transportChain: D: PDB Molecule: vacuolar protein-sorting-associated protein 25;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
22 c2zmeD_
not modelled
20.6
47
PDB header: protein transportChain: D: PDB Molecule: vacuolar protein-sorting-associated protein 25;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
23 d1baia_
not modelled
15.8
33
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)24 c2k8fB_
not modelled
14.7
29
PDB header: transferase/transcriptionChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: structural basis for the regulation of p53 function by p300
25 c3a2aC_
not modelled
11.6
69
PDB header: transport proteinChain: C: PDB Molecule: voltage-gated hydrogen channel 1;PDBTitle: the structure of the carboxyl-terminal domain of the human voltage-2 gated proton channel hv1
26 c3e0jG_
not modelled
11.5
26
PDB header: transferaseChain: G: PDB Molecule: dna polymerase subunit delta-2;PDBTitle: x-ray structure of the complex of regulatory subunits of2 human dna polymerase delta
27 c2zkrl_
not modelled
10.8
16
PDB header: ribosomal protein/rnaChain: L: PDB Molecule: rna expansion segment es20;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
28 c3bv6D_
not modelled
10.5
11
PDB header: hydrolaseChain: D: PDB Molecule: metal-dependent hydrolase;PDBTitle: crystal structure of uncharacterized metallo protein from vibrio2 cholerae with beta-lactamase like fold
29 d1sgua_
not modelled
9.8
20
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)30 d1v6fa_
not modelled
9.3
16
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Cofilin-like31 d1az5a_
not modelled
9.3
20
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)32 d1nvpd2
not modelled
9.2
38
Fold: Transcription factor IIA (TFIIA), beta-barrel domainSuperfamily: Transcription factor IIA (TFIIA), beta-barrel domainFamily: Transcription factor IIA (TFIIA), beta-barrel domain33 d1gkra1
not modelled
9.1
25
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)34 d2rspa_
not modelled
9.1
29
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)35 d1lyva_
not modelled
8.3
33
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases36 d1hvca_
not modelled
7.4
30
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)37 d2jdid2
not modelled
7.0
16
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase38 d2gu3a2
not modelled
7.0
16
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: PepSY-like39 d1mabb2
not modelled
6.8
16
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase40 d1nh2d2
not modelled
6.5
25
Fold: Transcription factor IIA (TFIIA), beta-barrel domainSuperfamily: Transcription factor IIA (TFIIA), beta-barrel domainFamily: Transcription factor IIA (TFIIA), beta-barrel domain41 c2gu3A_
not modelled
6.4
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ypmb protein;PDBTitle: ypmb protein from bacillus subtilis
42 d2bg1a1
not modelled
6.4
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase43 d1y7ea2
not modelled
6.2
12
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases44 c3lk3T_
not modelled
6.1
38
PDB header: protein bindingChain: T: PDB Molecule: leucine-rich repeat-containing protein 16a;PDBTitle: crystal structure of capz bound to the cpi and csi uncapping2 motifs from carmil
45 d3c7bb2
not modelled
5.9
36
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like46 d2c5wb1
not modelled
5.8
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase47 c2j8oA_
not modelled
5.8
11
PDB header: structural proteinChain: A: PDB Molecule: titin;PDBTitle: structure of the immunoglobulin tandem repeat of titin a168-2 a169
48 c3gebC_
not modelled
5.6
18
PDB header: hydrolaseChain: C: PDB Molecule: eyes absent homolog 2;PDBTitle: crystal structure of edeya2
49 c1tiiC_
not modelled
5.5
44
PDB header: enterotoxinChain: C: PDB Molecule: heat labile enterotoxin type iib;PDBTitle: escherichia coli heat labile enterotoxin type iib
50 d1nfga1
not modelled
5.5
30
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)51 d2gycj1
not modelled
5.4
27
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e52 c3c7bE_
not modelled
5.3
40
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
53 d1flga_
not modelled
5.2
28
Fold: 8-bladed beta-propellerSuperfamily: Quinoprotein alcohol dehydrogenase-likeFamily: Quinoprotein alcohol dehydrogenase-like54 d4fiva_
not modelled
5.2
15
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)55 c3t38B_
not modelled
5.1
15
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'