| 1 | c3louB_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 2 | c3o1lB_
|
|
 |
100.0 |
40 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 3 | c3obiC_
|
|
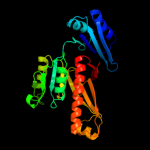 |
100.0 |
37 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 4 | c3n0vD_
|
|
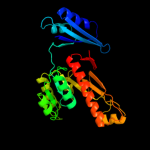 |
100.0 |
38 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 5 | c3nrbD_
|
|
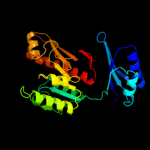 |
100.0 |
39 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 6 | c2ywrA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex2 aeolicus
|
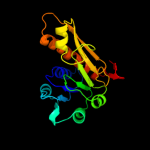
| 7 | d1jkxa_
|
|
 |
100.0 |
26 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
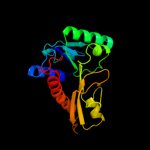
| 8 | c3p9xB_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
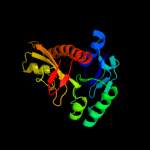
| 9 | c3tqrA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
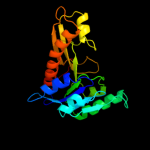
| 10 | c3dcjA_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide
PDBTitle: crystal structure of glycinamide formyltransferase (purn)2 from mycobacterium tuberculosis in complex with 5-methyl-5,3 6,7,8-tetrahydrofolic acid derivative
|
| 11 | d1meoa_
|
|
 |
100.0 |
29 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 12 | c3kcqA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
| 13 | d1fmta2
|
|
 |
100.0 |
21 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 14 | d2blna2
|
|
 |
100.0 |
21 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 15 | c1z7eC_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
| 16 | c1fmtA_
|
|
 |
100.0 |
19 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
| 17 | d1s3ia2
|
|
 |
100.0 |
18 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 18 | c3tqqA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: structure of the methionyl-trna formyltransferase (fmt) from coxiella2 burnetii
|
| 19 | c1yrwA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
| 20 | d2bw0a2
|
|
 |
100.0 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 21 | c3q0iA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from vibrio cholerae
|
| 22 | c3rfoA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
| 23 | c1s3iA_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: crystal structure of the n terminal hydrolase domain of 10-2 formyltetrahydrofolate dehydrogenase
|
| 24 | d1zgha2 |
|
not modelled |
100.0 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 25 | c1zghA_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from clostridium thermocellum
|
| 26 | d1zpva1 |
|
not modelled |
99.4 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 27 | c1u8sB_ |
|
not modelled |
99.3 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
| 28 | d1u8sa1 |
|
not modelled |
99.0 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 29 | c2nyiB_ |
|
not modelled |
99.0 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 30 | d1u8sa2 |
|
not modelled |
98.8 |
9 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 31 | c3p96A_ |
|
not modelled |
97.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase serb;
PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
|
| 32 | c1y7pB_ |
|
not modelled |
97.4 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein af1403;
PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
|
| 33 | c3ibwA_ |
|
not modelled |
97.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
| 34 | d1ygya3 |
|
not modelled |
97.1 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 35 | d1sc6a3 |
|
not modelled |
96.9 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 36 | c1ygyA_ |
|
not modelled |
96.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
|
| 37 | d2fgca2 |
|
not modelled |
96.2 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 38 | c2fgcA_ |
|
not modelled |
95.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
| 39 | c3a14B_ |
|
not modelled |
94.7 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
|
| 40 | d1ru8a_ |
|
not modelled |
94.7 |
10 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 41 | c2f1fA_ |
|
not modelled |
94.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
| 42 | d2pc6a2 |
|
not modelled |
93.9 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 43 | d2d13a1 |
|
not modelled |
93.8 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 44 | d1f0ka_ |
|
not modelled |
93.7 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 45 | c2pc6C_ |
|
not modelled |
93.2 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 46 | d1xi8a3 |
|
not modelled |
92.9 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 47 | c2e21A_ |
|
not modelled |
92.6 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
|
| 48 | c2hmaA_ |
|
not modelled |
92.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:probable trna (5-methylaminomethyl-2-thiouridylate)-
PDBTitle: the crystal structure of trna (5-methylaminomethyl-2-thiouridylate)-2 methyltransferase trmu from streptococcus pneumoniae
|
| 49 | d2f06a1 |
|
not modelled |
92.5 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 50 | d2f1fa1 |
|
not modelled |
92.5 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 51 | c2derA_ |
|
not modelled |
91.0 |
12 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna-specific 2-thiouridylase mnma;
PDBTitle: cocrystal structure of an rna sulfuration enzyme mnma and2 trna-glu in the initial trna binding state
|
| 52 | d1wy5a1 |
|
not modelled |
90.5 |
9 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 53 | d1r0ka2 |
|
not modelled |
90.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 54 | c1ni5A_ |
|
not modelled |
90.5 |
15 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative cell cycle protein mesj;
PDBTitle: structure of the mesj pp-atpase from escherichia coli
|
| 55 | c2jcyA_ |
|
not modelled |
90.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium3 tuberculosis
|
| 56 | c3bl5E_ |
|
not modelled |
90.0 |
9 |
PDB header:hydrolase
Chain: E: PDB Molecule:queuosine biosynthesis protein quec;
PDBTitle: crystal structure of quec from bacillus subtilis: an enzyme2 involved in preq1 biosynthesis
|
| 57 | d1ni5a1 |
|
not modelled |
89.9 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 58 | c2fu3A_ |
|
not modelled |
89.6 |
15 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
| 59 | d2ftsa3 |
|
not modelled |
89.5 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 60 | c3l76B_ |
|
not modelled |
88.5 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
| 61 | d1wu2a3 |
|
not modelled |
88.3 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 62 | d2f06a2 |
|
not modelled |
87.4 |
14 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 63 | c3a2kB_ |
|
not modelled |
86.8 |
13 |
PDB header:ligase/rna
Chain: B: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils complexed with trna
|
| 64 | c1ofgF_ |
|
not modelled |
86.7 |
10 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 65 | d1q0qa2 |
|
not modelled |
86.6 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 66 | c1h6dL_ |
|
not modelled |
86.5 |
10 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 67 | c2nvwB_ |
|
not modelled |
86.5 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 68 | c2nqqA_ |
|
not modelled |
86.2 |
16 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
| 69 | d1phza1 |
|
not modelled |
86.0 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 70 | c2q8nB_ |
|
not modelled |
85.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of glucose-6-phosphate isomerase (ec2 5.3.1.9) (tm1385) from thermotoga maritima at 1.82 a3 resolution
|
| 71 | c2qmxB_ |
|
not modelled |
85.6 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
|
| 72 | c3mwbA_ |
|
not modelled |
85.3 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
|
| 73 | c3luyA_ |
|
not modelled |
84.7 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable chorismate mutase;
PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
|
| 74 | d1jsca3 |
|
not modelled |
83.9 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 75 | d1uz5a3 |
|
not modelled |
83.5 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 76 | c3e18A_ |
|
not modelled |
83.3 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 77 | c3q2kB_ |
|
not modelled |
82.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 78 | c1uz5A_ |
|
not modelled |
82.4 |
11 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
| 79 | c3mtjA_ |
|
not modelled |
81.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
|
| 80 | c2f06B_ |
|
not modelled |
80.0 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
| 81 | c3lq1A_ |
|
not modelled |
80.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-
PDBTitle: crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene2 1-carboxylic acid synthase/2-oxoglutarate decarboxylase3 from listeria monocytogenes str. 4b f2365
|
| 82 | c2o48X_ |
|
not modelled |
79.4 |
13 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 83 | c3ceaA_ |
|
not modelled |
78.6 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 84 | c2pjkA_ |
|
not modelled |
77.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
| 85 | d2hmfa3 |
|
not modelled |
77.0 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 86 | d1ozha3 |
|
not modelled |
76.9 |
17 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 87 | c3e9mC_ |
|
not modelled |
75.8 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 88 | c2is8A_ |
|
not modelled |
75.5 |
8 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 89 | d1y5ea1 |
|
not modelled |
74.9 |
12 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 90 | d2f7wa1 |
|
not modelled |
74.9 |
10 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 91 | c3gd5D_ |
|
not modelled |
74.4 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
| 92 | c2vxoB_ |
|
not modelled |
73.9 |
9 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: human gmp synthetase in complex with xmp
|
| 93 | d1ybha3 |
|
not modelled |
73.6 |
16 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 94 | d1vh3a_ |
|
not modelled |
73.4 |
11 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 95 | d1h6da1 |
|
not modelled |
72.3 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 96 | c3ketA_ |
|
not modelled |
72.1 |
13 |
PDB header:transcription/dna
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
|
| 97 | c1xeaD_ |
|
not modelled |
71.9 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 98 | d1jlja_ |
|
not modelled |
71.8 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 99 | c3moiA_ |
|
not modelled |
71.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 100 | d1tq8a_ |
|
not modelled |
71.0 |
11 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 101 | c3db2C_ |
|
not modelled |
70.8 |
4 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 102 | c1evjC_ |
|
not modelled |
69.8 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: crystal structure of glucose-fructose oxidoreductase (gfor)2 delta1-22 s64d
|
| 103 | c1zh8B_ |
|
not modelled |
69.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 104 | d2hmfa2 |
|
not modelled |
69.4 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 105 | d3bula2 |
|
not modelled |
69.2 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 106 | d2djia3 |
|
not modelled |
68.8 |
18 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 107 | d1mvla_ |
|
not modelled |
68.6 |
14 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 108 | c1mvlA_ |
|
not modelled |
68.6 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:ppc decarboxylase athal3a;
PDBTitle: ppc decarboxylase mutant c175s
|
| 109 | d1vl2a1 |
|
not modelled |
68.5 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 110 | c1jscA_ |
|
not modelled |
68.3 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:acetohydroxy-acid synthase;
PDBTitle: crystal structure of the catalytic subunit of yeast2 acetohydroxyacid synthase: a target for herbicidal3 inhibitors
|
| 111 | d2qmwa2 |
|
not modelled |
68.0 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 112 | d2nqra3 |
|
not modelled |
66.6 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 113 | c2eghA_ |
|
not modelled |
65.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
|
| 114 | d1a9xa4 |
|
not modelled |
65.6 |
13 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 115 | c2ho3D_ |
|
not modelled |
65.6 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
| 116 | c2glxD_ |
|
not modelled |
62.8 |
10 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
| 117 | d2nvwa1 |
|
not modelled |
62.7 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 118 | d1q6za3 |
|
not modelled |
61.5 |
16 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 119 | c3rbvA_ |
|
not modelled |
61.1 |
19 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 120 | c2ag1A_ |
|
not modelled |
61.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:benzaldehyde lyase;
PDBTitle: crystal structure of benzaldehyde lyase (bal)- semet
|