| 1 |
|
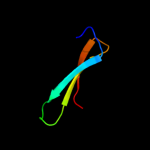
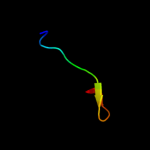
PDB 1fbv chain A
Region: 13 - 48
Aligned: 36
Modelled: 36
Confidence: 28.2%
Identity: 31%
PDB header:ligase
Chain: A: PDB Molecule:signal transduction protein cbl;
PDBTitle: structure of a cbl-ubch7 complex: ring domain function in2 ubiquitin-protein ligases
Phyre2
| 2 |
|
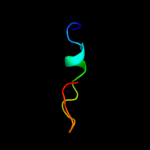
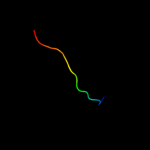
PDB 3bux chain B domain 3
Region: 13 - 47
Aligned: 35
Modelled: 35
Confidence: 27.1%
Identity: 31%
Fold: SH2-like
Superfamily: SH2 domain
Family: SH2 domain
Phyre2
| 3 |
|
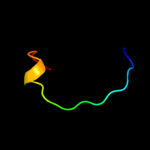
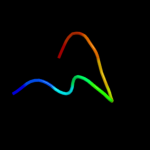
PDB 2cbl chain A
Region: 13 - 47
Aligned: 35
Modelled: 35
Confidence: 25.6%
Identity: 31%
PDB header:complex (proto-oncogene/peptide)
Chain: A: PDB Molecule:proto-oncogene cbl;
PDBTitle: n-terminal domain of cbl in complex with its binding site2 on zap-70
Phyre2
| 4 |
|
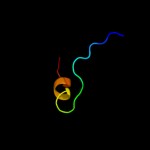
PDB 3bun chain B
Region: 13 - 47
Aligned: 35
Modelled: 35
Confidence: 25.1%
Identity: 31%
PDB header:ligase/signaling protein
Chain: B: PDB Molecule:e3 ubiquitin-protein ligase cbl;
PDBTitle: crystal structure of c-cbl-tkb domain complexed with its2 binding motif in sprouty4
Phyre2
| 5 |
|
PDB 1bnl chain A
Region: 22 - 34
Aligned: 13
Modelled: 13
Confidence: 15.6%
Identity: 38%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Endostatin
Phyre2
| 6 |
|
PDB 2gyr chain B
Region: 16 - 28
Aligned: 13
Modelled: 13
Confidence: 11.5%
Identity: 38%
PDB header:hormone/growth factor
Chain: B: PDB Molecule:neurotrophic factor artemin, isoform 3;
PDBTitle: crystal structure of human artemin
Phyre2
| 7 |
|
PDB 1koe chain A
Region: 22 - 34
Aligned: 13
Modelled: 13
Confidence: 10.5%
Identity: 38%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Endostatin
Phyre2
| 8 |
|
PDB 2qh7 chain A
Region: 19 - 27
Aligned: 9
Modelled: 9
Confidence: 8.6%
Identity: 44%
PDB header:metal binding protein
Chain: A: PDB Molecule:zinc finger cdgsh-type domain 1;
PDBTitle: mitoneet is a uniquely folded 2fe-2s outer mitochondrial membrane2 protein stabilized by pioglitazone
Phyre2
| 9 |
|
PDB 3fnv chain B
Region: 19 - 27
Aligned: 9
Modelled: 9
Confidence: 8.5%
Identity: 44%
PDB header:metal binding protein
Chain: B: PDB Molecule:cdgsh iron sulfur domain-containing protein 2;
PDBTitle: crystal structure of miner1: the redox-active 2fe-2s protein causative2 in wolfram syndrome 2
Phyre2
| 10 |
|
PDB 2zkq chain L
Region: 40 - 49
Aligned: 10
Modelled: 10
Confidence: 8.0%
Identity: 60%
PDB header:ribosomal protein/rna
Chain: L: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 11 |
|
PDB 3op0 chain B
Region: 13 - 47
Aligned: 35
Modelled: 35
Confidence: 7.9%
Identity: 34%
PDB header:signaling protein/signaling protein regu
Chain: B: PDB Molecule:signal transduction protein cbl-c;
PDBTitle: crystal structure of cbl-c (cbl-3) tkb domain in complex with egfr2 py1069 peptide
Phyre2
| 12 |
|
PDB 2pe4 chain A
Region: 21 - 41
Aligned: 18
Modelled: 21
Confidence: 7.7%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronidase-1;
PDBTitle: structure of human hyaluronidase 1, a hyaluronan hydrolyzing enzyme2 involved in tumor growth and angiogenesis
Phyre2
| 13 |
|
PDB 2uub chain L domain 1
Region: 30 - 49
Aligned: 19
Modelled: 20
Confidence: 7.4%
Identity: 47%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 14 |
|
PDB 2xzm chain L
Region: 40 - 49
Aligned: 10
Modelled: 10
Confidence: 7.3%
Identity: 60%
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s12;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 15 |
|
PDB 2ilx chain A domain 1
Region: 22 - 43
Aligned: 22
Modelled: 22
Confidence: 6.7%
Identity: 32%
Fold: LigT-like
Superfamily: LigT-like
Family: 2',3'-cyclic nucleotide 3'-phosphodiesterase, catalytic domain
Phyre2
| 16 |
|
PDB 2zom chain C
Region: 21 - 36
Aligned: 16
Modelled: 16
Confidence: 6.5%
Identity: 31%
PDB header:unknown function
Chain: C: PDB Molecule:protein cuta, chloroplast, putative, expressed;
PDBTitle: crystal structure of cuta1 from oryza sativa
Phyre2
| 17 |
|
PDB 1agq chain A
Region: 16 - 28
Aligned: 13
Modelled: 13
Confidence: 6.5%
Identity: 54%
Fold: Cystine-knot cytokines
Superfamily: Cystine-knot cytokines
Family: Transforming growth factor (TGF)-beta
Phyre2
| 18 |
|
PDB 1s1h chain L
Region: 40 - 49
Aligned: 10
Modelled: 10
Confidence: 6.5%
Identity: 50%
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s23;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
Phyre2
| 19 |
|
PDB 1ok8 chain A domain 1
Region: 22 - 32
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 36%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: Class II viral fusion proteins C-terminal domain
Phyre2
| 20 |
|
PDB 3egp chain A
Region: 22 - 32
Aligned: 11
Modelled: 11
Confidence: 5.7%
Identity: 27%
PDB header:viral protein
Chain: A: PDB Molecule:envelope protein;
PDBTitle: crystal structure analysis of dengue-1 envelope protein2 domain iii
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|