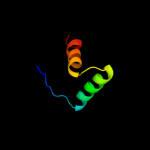
1 d1mnta_
89.7
30
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors2 c1u9pA_
79.8
22
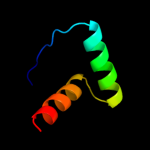
PDB header: unknown functionChain: A: PDB Molecule: parc;PDBTitle: permuted single-chain arc
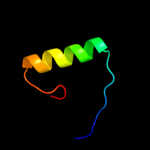
3 d1myla_
53.5
23
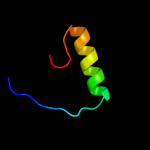
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors4 c2kkeA_
51.9
39
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a dimeric protein of unknown2 function from methanobacterium thermoautotrophicum,3 northeast structural genomics consortium target tr5
5 d1b28a_
51.8
23
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors6 d1mylb_
50.4
23
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors7 d1bazb_
47.6
28
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors8 d1baza_
45.4
28
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors9 d1bdta_
44.8
28
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors10 d1myka_
44.5
30
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors11 c3gtdB_
32.9
17
PDB header: lyaseChain: B: PDB Molecule: fumarate hydratase class ii;PDBTitle: 2.4 angstrom crystal structure of fumarate hydratase from rickettsia2 prowazekii
12 c3ns4A_
31.3
16
PDB header: protein bindingChain: A: PDB Molecule: vacuolar protein sorting-associated protein 53;PDBTitle: structure of a c-terminal fragment of its vps53 subunit suggests2 similarity of garp to a family of tethering complexes
13 d1c5ea_
29.9
35
Fold: beta-clipSuperfamily: Head decoration protein D (gpD, major capsid protein D)Family: Head decoration protein D (gpD, major capsid protein D)14 c2dvlB_
27.1
25
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of project tt0160 from thermus thermophilus hb8
15 c3h37B_
26.9
17
PDB header: transferaseChain: B: PDB Molecule: trna nucleotidyl transferase-related protein;PDBTitle: the structure of cca-adding enzyme apo form i
16 d1td4a_
24.2
29
Fold: beta-clipSuperfamily: Head decoration protein D (gpD, major capsid protein D)Family: Head decoration protein D (gpD, major capsid protein D)17 d1egda2
22.7
23
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains18 c2a1tC_
22.6
23
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: acyl-coa dehydrogenase, medium-chain specific,PDBTitle: structure of the human mcad:etf e165betaa complex
19 c1egcB_
19.2
23
PDB header: electron transferChain: B: PDB Molecule: medium chain acyl-coa dehydrogenase;PDBTitle: structure of t255e, e376g mutant of human medium chain acyl-2 coa dehydrogenase complexed with octanoyl-coa
20 c1ukwA_
18.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of medium-chain acyl-coa dehydrogenase2 from thermus thermophilus hb8
21 c2pg0B_
not modelled
18.1
21
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from geobacillus2 kaustophilus
22 d1grja1
not modelled
17.7
30
Fold: Long alpha-hairpinSuperfamily: GreA transcript cleavage protein, N-terminal domainFamily: GreA transcript cleavage protein, N-terminal domain23 c3nf4B_
not modelled
17.3
17
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from mycobacterium2 thermoresistibile bound to flavin adenine dinucleotide
24 c2cx9C_
not modelled
17.1
12
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase
25 c2derA_
not modelled
16.5
28
PDB header: transferase/rnaChain: A: PDB Molecule: trna-specific 2-thiouridylase mnma;PDBTitle: cocrystal structure of an rna sulfuration enzyme mnma and2 trna-glu in the initial trna binding state
26 c2e7vA_
not modelled
15.5
11
PDB header: hydrolaseChain: A: PDB Molecule: transmembrane protease;PDBTitle: crystal structure of sea domain of transmembrane protease2 from mus musculus
27 d2i09a2
not modelled
14.8
20
Fold: DeoB insert domain-likeSuperfamily: DeoB insert domain-likeFamily: DeoB insert domain-like28 d3mdea2
not modelled
14.1
22
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains29 d2pb9a1
not modelled
14.1
32
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: Phosphomethylpyrimidine kinase C-terminal domain-like30 d1pp7u_
not modelled
13.0
46
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: 39 kda initiator binding protein, IBP39, N-terminal domain31 c2rp4C_
not modelled
12.2
50
PDB header: transcriptionChain: C: PDB Molecule: transcription factor p53;PDBTitle: solution structure of the oligomerization domain in dmp53
32 c2jvwA_
not modelled
12.1
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of uncharacterized protein q5e7h1 from vibrio2 fischeri. northeast structural genomics target vfr117
33 c2a5wC_
not modelled
12.1
24
PDB header: oxidoreductaseChain: C: PDB Molecule: sulfite reductase, desulfoviridin-type subunit gammaPDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
34 d1go3f_
not modelled
12.0
30
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: RNA polymerase II subunit RBP4 (RpoF)35 c1tt9B_
not modelled
10.9
29
PDB header: transferase, lyaseChain: B: PDB Molecule: formimidoyltransferase-cyclodeaminasePDBTitle: structure of the bifunctional and golgi associated2 formiminotransferase cyclodeaminase octamer
36 c2kgnA_
not modelled
10.7
55
PDB header: signaling proteinChain: A: PDB Molecule: protein ste5;PDBTitle: solution structure of ste5pm24 in the zwitterionic dpc2 micelle
37 c2vigC_
not modelled
10.3
21
PDB header: oxidoreductaseChain: C: PDB Molecule: short-chain specific acyl-coa dehydrogenase,;PDBTitle: crystal structure of human short-chain acyl coa2 dehydrogenase
38 c2pjhA_
not modelled
10.3
19
PDB header: transport proteinChain: A: PDB Molecule: nuclear protein localization protein 4 homolog;PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
39 c1rx0B_
not modelled
10.1
14
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase family member 8,PDBTitle: crystal structure of isobutyryl-coa dehydrogenase complexed2 with substrate/ligand.
40 c3ghfA_
not modelled
9.5
18
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
41 c3bjqA_
not modelled
9.2
29
PDB header: viral proteinChain: A: PDB Molecule: phage-related protein;PDBTitle: crystal structure of a phage-related protein (bb3626) from bordetella2 bronchiseptica rb50 at 2.05 a resolution
42 c2l4uA_
not modelled
8.9
55
PDB header: signaling proteinChain: A: PDB Molecule: 24mer peptide from protein ste5;PDBTitle: solution structure of ste5pm24 in the presence of sds micelle
43 d2phpa1
not modelled
8.9
29
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: Phosphomethylpyrimidine kinase C-terminal domain-like44 c2jifA_
not modelled
8.8
14
PDB header: oxidoreductaseChain: A: PDB Molecule: short/branched chain specific acyl-coa dehydrogenase;PDBTitle: structure of human short-branched chain acyl-coa2 dehydrogenase (acadsb)
45 c2kgmA_
not modelled
8.5
55
PDB header: signaling proteinChain: A: PDB Molecule: protein ste5;PDBTitle: solution structure of ste5pm24 in sds micelle
46 d1qyna_
not modelled
8.3
17
Fold: SecB-likeSuperfamily: SecB-likeFamily: Bacterial protein-export protein SecB47 c3hieA_
not modelled
8.3
45
PDB header: exocytosisChain: A: PDB Molecule: exocyst complex component sec3;PDBTitle: structure of the membrane-binding domain of the sec3 subunit2 of the exocyst complex
48 c3r7kB_
not modelled
8.3
18
PDB header: oxidoreductaseChain: B: PDB Molecule: probable acyl coa dehydrogenase;PDBTitle: crystal structure of a probable acyl coa dehydrogenase from2 mycobacterium abscessus atcc 19977 / dsm 44196
49 d1u5ea1
not modelled
8.2
14
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)50 c2zq5A_
not modelled
8.1
11
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of sulfotransferase stf1 from2 mycobacterium tuberculosis h37rv (type1 form)
51 c2gsjA_
not modelled
7.7
24
PDB header: hydrolaseChain: A: PDB Molecule: protein ppl-2;PDBTitle: cdna cloning and 1.75a crystal structure determination of2 ppl2, a novel chimerolectin from parkia platycephala seeds3 exhibiting endochitinolytic activity
52 c3t38B_
not modelled
7.6
32
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
53 c2kb2A_
not modelled
7.4
27
PDB header: signaling protein, hydrolase regulatorChain: A: PDB Molecule: blrp1;PDBTitle: blrp1 bluf
54 d1yrxa1
not modelled
7.2
27
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: BLUF domain55 d1jqia2
not modelled
7.0
18
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains56 d1v5ma_
not modelled
6.9
5
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)57 d2ccva1
not modelled
6.7
21
Fold: HPA-likeSuperfamily: Agglutinin HPA-likeFamily: Agglutinin HPA-like58 c2p0fA_
not modelled
6.6
23
PDB header: ligand binding proteinChain: A: PDB Molecule: rho gtpase-activating protein 9;PDBTitle: arhgap9 ph domain in complex with ins(1,3,5)p3
59 d1ciya3
not modelled
6.6
23
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain60 d1ukwa2
not modelled
6.5
23
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains61 c3oibB_
not modelled
6.4
25
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of a putative acyl-coa dehydrogenase from2 mycobacterium smegmatis, iodide soak
62 d1us3a2
not modelled
6.4
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases63 d1t3ta1
not modelled
6.3
32
Fold: RuvA C-terminal domain-likeSuperfamily: FGAM synthase PurL, linker domainFamily: FGAM synthase PurL, linker domain64 c1mpgB_
not modelled
6.1
30
PDB header: hydrolaseChain: B: PDB Molecule: 3-methyladenine dna glycosylase ii;PDBTitle: 3-methyladenine dna glycosylase ii from escherichia coli
65 d1o6da_
not modelled
6.0
42
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like66 c2fknC_
not modelled
5.8
19
PDB header: lyaseChain: C: PDB Molecule: urocanate hydratase;PDBTitle: crystal structure of urocanase from bacillus subtilis
67 c3a58A_
not modelled
5.8
45
PDB header: protein transport/exocytosisChain: A: PDB Molecule: exocyst complex component sec3;PDBTitle: crystal structure of sec3p - rho1p complex from2 saccharomyces cerevisiae
68 d1uwka_
not modelled
5.8
31
Fold: UrocanaseSuperfamily: UrocanaseFamily: Urocanase69 c2xqyA_
not modelled
5.8
39
PDB header: immune system/viral proteinChain: A: PDB Molecule: envelope glycoprotein h;PDBTitle: crystal structure of pseudorabies core fragment of2 glycoprotein h in complex with fab d6.3
70 d1qy9a1
not modelled
5.5
15
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like71 d1ofcx2
not modelled
5.4
40
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: SLIDE domain72 c3a9lB_
not modelled
5.3
27
PDB header: hydrolaseChain: B: PDB Molecule: poly-gamma-glutamate hydrolase;PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
73 d1vk3a1
not modelled
5.3
29
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like74 c1yx3A_
not modelled
5.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
75 c2ebaI_
not modelled
5.3
18
PDB header: oxidoreductaseChain: I: PDB Molecule: putative glutaryl-coa dehydrogenase;PDBTitle: crystal structure of the putative glutaryl-coa dehydrogenase from2 thermus thermophilus
76 d1miwa2
not modelled
5.1
22
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like77 d1droa_
not modelled
5.1
5
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)78 c2jyaA_
not modelled
5.1
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1810;PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
79 d1h4ra3
not modelled
5.0
16
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM