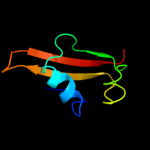
1 c2gu1A_
99.2
24
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
2 c2l9yA_
98.4
13
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
3 c2djpA_
98.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
4 d1e0ga_
98.3
31
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain5 d1y7ma2
98.1
23
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain6 c1y7mB_
97.2
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein bsu14040;PDBTitle: crystal structure of the b. subtilis ykud protein at 2 a2 resolution
7 c2l55A_
58.2
19
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
8 d1rr7a_
55.8
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor9 c1rr7A_
55.8
21
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
10 c1ei5A_
53.6
24
PDB header: hydrolaseChain: A: PDB Molecule: d-aminopeptidase;PDBTitle: crystal structure of a d-aminopeptidase from ochrobactrum2 anthropi
11 c1vzyA_
43.6
19
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of the bacillus subtilis hsp33
12 d1nera_
42.8
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors13 d2obpa1
42.7
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ReutB4095-like14 c1zeqX_
42.4
17
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
15 d2jn6a1
42.0
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Cgl2762-like16 c2elhA_
40.2
23
PDB header: dna binding proteinChain: A: PDB Molecule: cg11849-pa;PDBTitle: solution structure of the cenp-b n-terminal dna-binding2 domain of fruit fly distal antenna cg11849-pa
17 c2pmzV_
38.9
8
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
18 c1vq0A_
38.8
11
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of 33 kda chaperonin (heat shock protein 33 homolog)2 (hsp33) (tm1394) from thermotoga maritima at 2.20 a resolution
19 c2r0qF_
37.0
15
PDB header: recombination/dnaChain: F: PDB Molecule: putative transposon tn552 dna-invertase bin3;PDBTitle: crystal structure of a serine recombinase- dna regulatory2 complex
20 d1hmja_
35.5
8
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB521 d1je3a_
not modelled
35.2
13
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like22 d1vq0a1
not modelled
32.4
11
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain23 c1hw7A_
not modelled
31.8
21
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp33;PDBTitle: hsp33, heat shock protein with redox-regulated chaperone activity
24 d1hw7a_
not modelled
31.8
21
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain25 c3iswA_
not modelled
31.5
14
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: crystal structure of filamin-a immunoglobulin-like repeat 21 bound to2 an n-terminal peptide of cftr
26 d1eika_
not modelled
31.4
10
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB527 d1vzya1
not modelled
30.8
17
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain28 d1uxca_
not modelled
29.0
13
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator29 c1i7fA_
not modelled
28.9
20
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 33;PDBTitle: crystal structure of the hsp33 domain with constitutive chaperone2 activity
30 d2i6va1
not modelled
28.6
22
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like31 c3mmlE_
not modelled
27.8
29
PDB header: hydrolaseChain: E: PDB Molecule: allophanate hydrolase subunit 2;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
32 d2i4sa1
not modelled
25.0
22
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like33 c3cecA_
not modelled
24.6
21
PDB header: transcriptionChain: A: PDB Molecule: putative antidote protein of plasmid maintenance system;PDBTitle: crystal structure of a putative antidote protein of plasmid2 maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at3 1.60 a resolution
34 d1dzfa2
not modelled
24.6
18
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB535 d1dcja_
not modelled
24.5
13
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like36 c2ia2D_
not modelled
24.3
8
PDB header: transcriptionChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: the crystal structure of a putative transcriptional regulator rha061952 from rhodococcus sp. rha1
37 d1i7ha_
not modelled
23.8
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related38 c3hefB_
not modelled
23.5
10
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
39 d2fug33
not modelled
22.8
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins40 d1hfua2
not modelled
22.7
12
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins41 d1twfc2
not modelled
22.6
29
Fold: Insert subdomain of RNA polymerase alpha subunitSuperfamily: Insert subdomain of RNA polymerase alpha subunitFamily: Insert subdomain of RNA polymerase alpha subunit42 d1uxda_
not modelled
22.5
13
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator43 c2zpmA_
not modelled
22.4
6
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
44 d3c8ya2
not modelled
22.4
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins45 c2gm4B_
not modelled
22.2
12
PDB header: recombination, dnaChain: B: PDB Molecule: transposon gamma-delta resolvase;PDBTitle: an activated, tetrameric gamma-delta resolvase: hin chimaera bound to2 cleaved dna
46 d2proc1
not modelled
22.2
50
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain47 d1y0pa3
not modelled
21.9
8
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain48 d1d1la_
not modelled
21.9
22
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors49 c2pijB_
not modelled
21.2
22
PDB header: transcriptionChain: B: PDB Molecule: prophage pfl 6 cro;PDBTitle: structure of the cro protein from prophage pfl 6 in pseudomonas2 fluorescens pf-5
50 d2q9oa2
not modelled
21.2
12
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins51 c2rn7A_
not modelled
21.1
9
PDB header: unknown functionChain: A: PDB Molecule: is629 orfa;PDBTitle: nmr solution structure of tnpe protein from shigella2 flexneri. northeast structural genomics target sfr125
52 d1wlha1
not modelled
20.9
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)53 c2brqB_
not modelled
20.9
14
PDB header: structural proteinChain: B: PDB Molecule: filamin a;PDBTitle: crystal structure of the filamin a repeat 21 complexed with2 the integrin beta7 cytoplasmic tail peptide
54 d1ei5a2
not modelled
20.9
24
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains55 c2lcvA_
not modelled
20.7
13
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
56 d1v10a2
not modelled
20.3
12
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins57 d1fdra1
not modelled
20.1
29
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like58 c3fiaA_
not modelled
20.1
16
PDB header: protein bindingChain: A: PDB Molecule: intersectin-1;PDBTitle: crystal structure of the eh 1 domain from human intersectin-2 1 protein. northeast structural genomics consortium target3 hr3646e.
59 d1kyaa2
not modelled
19.9
10
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins60 c2gu3A_
not modelled
19.0
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ypmb protein;PDBTitle: ypmb protein from bacillus subtilis
61 c3f6vA_
not modelled
18.6
13
PDB header: transcription regulatorChain: A: PDB Molecule: possible transcriptional regulator, arsr familyPDBTitle: crystal structure of possible transcriptional regulator for2 arsenical resistance
62 d4fxca_
not modelled
18.3
7
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related63 c1p0oA_
not modelled
18.3
25
PDB header: ribosomeChain: A: PDB Molecule: 19-mer peptide from 50s ribosomal protein l1;PDBTitle: hp (2-20) substitution of trp for gln and asp at position2 17 and 19 modification in sds-d25 micelles
64 d1aoza2
not modelled
18.1
20
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins65 d1a8pa1
not modelled
18.1
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like66 d1gyca2
not modelled
17.8
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins67 c2l8nA_
not modelled
17.7
12
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional repressor cytr;PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
68 c3lxfC_
not modelled
17.2
28
PDB header: metal binding proteinChain: C: PDB Molecule: ferredoxin;PDBTitle: crystal structure of [2fe-2s] ferredoxin arx from novosphingobium2 aromaticivorans
69 d2gu3a1
not modelled
16.8
10
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: PepSY-like70 d4croa_
not modelled
16.7
22
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors71 d1awda_
not modelled
16.5
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related72 d1bw6a_
not modelled
16.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding73 d1hlva1
not modelled
16.4
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding74 d2cuaa_
not modelled
16.0
20
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Periplasmic domain of cytochrome c oxidase subunit II75 d2hsga1
not modelled
15.8
13
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator76 c3cuoB_
not modelled
15.6
17
PDB header: transcription regulatorChain: B: PDB Molecule: uncharacterized hth-type transcriptional regulator ygav;PDBTitle: crystal structure of the predicted dna-binding transcriptional2 regulator from e. coli
77 c2kjpA_
not modelled
15.6
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ylbl;PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
78 c2xroE_
not modelled
15.3
16
PDB header: dna-binding protein/dnaChain: E: PDB Molecule: hth-type transcriptional regulator ttgv;PDBTitle: crystal structure of ttgv in complex with its dna operator
79 d1mkma1
not modelled
15.1
4
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator IclR, N-terminal domain80 c3h0gE_
not modelled
15.0
15
PDB header: transcriptionChain: E: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
81 d2jxca1
not modelled
15.0
10
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)82 d2icta1
not modelled
15.0
7
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like83 d1nxza1
not modelled
14.9
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like84 d1zud21
not modelled
14.7
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS85 d1lcda_
not modelled
14.7
12
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator86 c3c19A_
not modelled
14.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mk0293;PDBTitle: crystal structure of protein mk0293 from methanopyrus kandleri av19
87 d1jiwi_
not modelled
14.4
19
Fold: Streptavidin-likeSuperfamily: beta-Barrel protease inhibitorsFamily: Metalloprotease inhibitor88 c1mkmA_
not modelled
14.3
4
PDB header: transcriptionChain: A: PDB Molecule: iclr transcriptional regulator;PDBTitle: crystal structure of the thermotoga maritima iclr
89 d1wxma1
not modelled
13.9
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD90 c2otgA_
not modelled
13.9
19
PDB header: contractile proteinChain: A: PDB Molecule: myosin heavy chain;PDBTitle: rigor-like structures of muscle myosins reveal key2 mechanical elements in the transduction pathways of this3 allosteric motor
91 c3p42D_
not modelled
13.7
25
PDB header: unknown functionChain: D: PDB Molecule: predicted protein;PDBTitle: structure of gfcc (ymcb), protein encoded by the e. coli group 42 capsule operon
92 d1e9ma_
not modelled
13.6
28
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related93 d3orca_
not modelled
13.4
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors94 d1hh2p4
not modelled
13.3
18
Fold: Transcription factor NusA, N-terminal domainSuperfamily: Transcription factor NusA, N-terminal domainFamily: Transcription factor NusA, N-terminal domain95 c2kfhA_
not modelled
13.3
16
PDB header: protein bindingChain: A: PDB Molecule: eh domain-containing protein 1;PDBTitle: structure of the c-terminal domain of ehd1 with fnyestgpftak
96 d1kk8a2
not modelled
13.2
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins97 d1d0xa2
not modelled
13.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins98 c2o0yB_
not modelled
13.1
19
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator rha1_ro069532 (iclr-family) from rhodococcus sp.
99 d1d4ca3
not modelled
13.1
27
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain