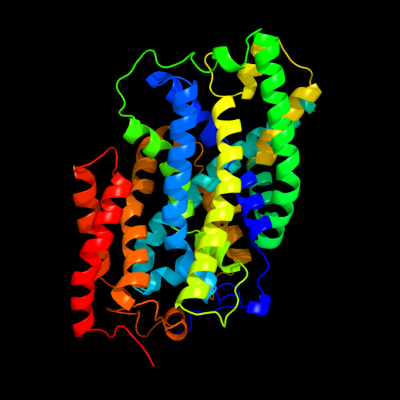
| 1 | c2jlnA_
|
|
 |
100.0 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
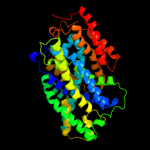
| 2 | c2xq2A_
|
|
 |
99.7 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
|
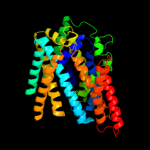
| 3 | c3dh4A_
|
|
 |
99.6 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
|
| 4 | c3giaA_
|
|
 |
99.4 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
|
| 5 | c3lrcC_
|
|
 |
99.2 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
|
| 6 | d2a65a1
|
|
 |
97.2 |
10 |
Fold:SNF-like
Superfamily:SNF-like
Family:SNF-like |
| 7 | c2w8aC_
|
|
 |
96.4 |
12 |
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
|
| 8 | c3hfxA_
|
|
 |
96.1 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
|
| 9 | d1pw4a_
|
|
 |
46.3 |
10 |
Fold:MFS general substrate transporter
Superfamily:MFS general substrate transporter
Family:Glycerol-3-phosphate transporter |
| 10 | c2bg9C_
|
|
 |
37.7 |
12 |
PDB header:ion channel/receptor
Chain: C: PDB Molecule:acetylcholine receptor protein, delta chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
|
| 11 | c3qnqD_
|
|
 |
27.7 |
30 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
| 12 | c2bg9A_
|
|
 |
23.4 |
25 |
PDB header:ion channel/receptor
Chain: A: PDB Molecule:acetylcholine receptor protein, alpha chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
|
| 13 | c3m7bA_
|
|
 |
21.0 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
|
| 14 | c2qc1B_
|
|
 |
14.5 |
25 |
PDB header:protein binding
Chain: B: PDB Molecule:acetylcholine receptor subunit alpha;
PDBTitle: crystal structure of the extracellular domain of the nicotinic2 acetylcholine receptor 1 subunit bound to alpha-bungarotoxin at 1.9 a3 resolution
|
| 15 | d1iwga8
|
|
 |
14.2 |
7 |
Fold:Multidrug efflux transporter AcrB transmembrane domain
Superfamily:Multidrug efflux transporter AcrB transmembrane domain
Family:Multidrug efflux transporter AcrB transmembrane domain |
| 16 | c2bg9E_
|
|
 |
13.8 |
13 |
PDB header:ion channel/receptor
Chain: E: PDB Molecule:acetylcholine receptor protein, gamma chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
|
| 17 | c1r8jB_
|
|
 |
9.4 |
32 |
PDB header:circadian clock protein
Chain: B: PDB Molecule:kaia;
PDBTitle: crystal structure of circadian clock protein kaia from2 synechococcus elongatus
|
| 18 | c2raxY_
|
|
 |
9.2 |
24 |
PDB header:cell cycle
Chain: Y: PDB Molecule:borealin;
PDBTitle: crystal structure of borealin (20-78) bound to survivin (1-120)
|
| 19 | d1nj8a2
|
|
 |
7.6 |
31 |
Fold:IF3-like
Superfamily:C-terminal domain of ProRS
Family:C-terminal domain of ProRS |
| 20 | c3bowC_
|
|
 |
7.4 |
32 |
PDB header:hydrolase/hydrolase inhibitor
Chain: C: PDB Molecule:calpastatin;
PDBTitle: structure of m-calpain in complex with calpastatin
|
| 21 | d2auwa1 |
|
not modelled |
6.8 |
13 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:NE0471 C-terminal domain-like |
| 22 | d1eexa_ |
|
not modelled |
6.6 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Diol dehydratase, alpha subunit |
| 23 | d1t98a1 |
|
not modelled |
6.0 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MukF N-terminal domain-like |
| 24 | c1nauA_ |
|
not modelled |
5.9 |
32 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:glucagon;
PDBTitle: nmr solution structure of the glucagon antagonist [deshis1,2 desphe6, glu9]glucagon amide in the presence of3 perdeuterated dodecylphosphocholine micelles
|
| 25 | c2auwB_ |
|
not modelled |
5.5 |
13 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ne0471;
PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
|
| 26 | d1zbxb1 |
|
not modelled |
5.4 |
24 |
Fold:ORC1-binding domain
Superfamily:ORC1-binding domain
Family:ORC1-binding domain |
| 27 | c2k2wA_ |
|
not modelled |
5.4 |
11 |
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
|
| 28 | c2wj8N_ |
|
not modelled |
5.3 |
7 |
PDB header:rna binding protein/rna
Chain: N: PDB Molecule:nucleoprotein;
PDBTitle: respiratory syncitial virus ribonucleoprotein
|
| 29 | d1e6va2 |
|
not modelled |
5.3 |
20 |
Fold:Ferredoxin-like
Superfamily:Methyl-coenzyme M reductase subunits
Family:Methyl-coenzyme M reductase alpha and beta chain N-terminal domain |
| 30 | c1bh0A_ |
|
not modelled |
5.3 |
27 |
PDB header:synthetic hormone
Chain: A: PDB Molecule:glucagon;
PDBTitle: structure of a glucagon analog
|
| 31 | d2fgca2 |
|
not modelled |
5.1 |
44 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |