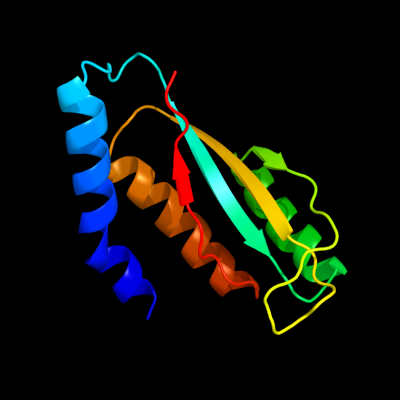
| 1 | d1nxia_
|
|
 |
100.0 |
65 |
Fold:Ferredoxin-like
Superfamily:Hypothetical protein VC0424
Family:Hypothetical protein VC0424 |
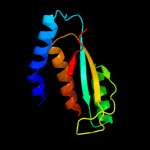
| 2 | d1j5ya2
|
|
 |
47.4 |
33 |
Fold:HPr-like
Superfamily:Putative transcriptional regulator TM1602, C-terminal domain
Family:Putative transcriptional regulator TM1602, C-terminal domain |
| 3 | c3tqxA_
|
|
 |
35.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-3-ketobutyrate coenzyme a ligase;
PDBTitle: structure of the 2-amino-3-ketobutyrate coenzyme a ligase (kbl) from2 coxiella burnetii
|
| 4 | d1fc4a_
|
|
 |
28.6 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 5 | c3hqtB_
|
|
 |
27.5 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:cai-1 autoinducer synthase;
PDBTitle: plp-dependent acyl-coa transferase cqsa
|
| 6 | c2p7pB_
|
|
 |
26.9 |
24 |
PDB header:metal binding protein, hydrolase
Chain: B: PDB Molecule:glyoxalase family protein;
PDBTitle: crystal structure of genomically encoded fosfomycin resistance2 protein, fosx, from listeria monocytogenes complexed with mn(ii) and3 sulfate ion
|
| 7 | c3qwuA_
|
|
 |
26.4 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:dna ligase;
PDBTitle: putative atp-dependent dna ligase from aquifex aeolicus.
|
| 8 | c3a2bA_
|
|
 |
25.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
|
| 9 | d2c0ra1
|
|
 |
24.7 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 10 | c2w8wA_
|
|
 |
22.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: n100y spt with plp-ser
|
| 11 | d2fhfa3
|
|
 |
20.5 |
18 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 12 | c2x48B_
|
|
 |
18.7 |
24 |
PDB header:viral protein
Chain: B: PDB Molecule:cag38821;
PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
|
| 13 | d2je6i2
|
|
 |
18.2 |
27 |
Fold:Barrel-sandwich hybrid
Superfamily:Ribosomal L27 protein-like
Family:ECR1 N-terminal domain-like |
| 14 | c3qm2A_
|
|
 |
16.8 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
|
| 15 | c3hz7A_
|
|
 |
16.3 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the sira-like protein (dsy4693) from2 desulfitobacterium hafniense, northeast structural genomics3 consortium target dhr2a
|
| 16 | d1o59a1
|
|
 |
16.3 |
83 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Allantoicase repeat |
| 17 | d1r9ca_
|
|
 |
15.8 |
19 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Antibiotic resistance proteins |
| 18 | c1zswA_
|
|
 |
14.8 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:glyoxalase family protein;
PDBTitle: crystal structure of bacillus cereus metallo protein from glyoxalase2 family
|
| 19 | c1z64A_
|
|
 |
14.6 |
67 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:pleruocidin;
PDBTitle: nmr solution structure of pleurocidin in dpc micelles
|
| 20 | d2j44a2
|
|
 |
14.5 |
18 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 21 | d2j43a1 |
|
not modelled |
13.8 |
9 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 22 | c1w1nA_ |
|
not modelled |
13.7 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol 3-kinase tor1;
PDBTitle: the solution structure of the fatc domain of the protein2 kinase tor1 from yeast
|
| 23 | c2qv5A_ |
|
not modelled |
13.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu2773;
PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
|
| 24 | c1j5yA_ |
|
not modelled |
12.5 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, biotin repressor family;
PDBTitle: crystal structure of transcriptional regulator (tm1602) from2 thermotoga maritima at 2.3 a resolution
|
| 25 | d2j73a1 |
|
not modelled |
12.4 |
45 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 26 | c2j43A_ |
|
not modelled |
12.2 |
18 |
PDB header:carbohydrate-binding module
Chain: A: PDB Molecule:spydx;
PDBTitle: alpha-glucan recognition by family 41 carbohydrate-binding2 modules from streptococcal virulence factors
|
| 27 | c2ke4A_ |
|
not modelled |
11.8 |
45 |
PDB header:membrane protein
Chain: A: PDB Molecule:cdc42-interacting protein 4;
PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
|
| 28 | d2j43a2 |
|
not modelled |
11.8 |
25 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 29 | c3m2oB_ |
|
not modelled |
11.7 |
36 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:glyoxalase/bleomycin resistance protein;
PDBTitle: crystal structure of a putative glyoxalase/bleomycin resistance2 protein from rhodopseudomonas palustris cga009
|
| 30 | c3dxvA_ |
|
not modelled |
11.6 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:alpha-amino-epsilon-caprolactam racemase;
PDBTitle: the crystal structure of alpha-amino-epsilon-caprolactam racemase from2 achromobacter obae
|
| 31 | d2j44a1 |
|
not modelled |
11.4 |
27 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 32 | d1utaa_ |
|
not modelled |
11.2 |
14 |
Fold:Ferredoxin-like
Superfamily:Sporulation related repeat
Family:Sporulation related repeat |
| 33 | c2j44A_ |
|
not modelled |
11.2 |
38 |
PDB header:carbohydrate-binding module
Chain: A: PDB Molecule:alkaline amylopullulanase;
PDBTitle: alpha-glucan binding by a streptococcal virulence factor
|
| 34 | d1mqta_ |
|
not modelled |
11.0 |
57 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 35 | d1bs0a_ |
|
not modelled |
10.7 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 36 | d1x9ba_ |
|
not modelled |
10.1 |
16 |
Fold:Protozoan pheromone-like
Superfamily:Hypothetical membrane protein Ta0354, soluble domain
Family:Hypothetical membrane protein Ta0354, soluble domain |
| 37 | c2f06B_ |
|
not modelled |
10.1 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
| 38 | d1oopa_ |
|
not modelled |
9.9 |
57 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 39 | c1z7s1_ |
|
not modelled |
9.7 |
26 |
PDB header:virus
Chain: 1: PDB Molecule:human coxsackievirus a21;
PDB Fragment:viral protein 1;
PDBTitle: the crystal structure of coxsackievirus a21
|
| 40 | c1wv9B_ |
|
not modelled |
8.9 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:rhodanese homolog tt1651;
PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
|
| 41 | c1zbe1_ |
|
not modelled |
8.7 |
57 |
PDB header:virus
Chain: 1: PDB Molecule:coat protein vp1;
PDBTitle: foot-and mouth disease virus serotype a1061
|
| 42 | d1o59a2 |
|
not modelled |
8.6 |
80 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Allantoicase repeat |
| 43 | c2wff1_ |
|
not modelled |
8.5 |
57 |
PDB header:virus
Chain: 1: PDB Molecule:p1;
PDB Fragment:capsid protein vp4, residues 1-80
PDBTitle: equine rhinitis a virus
|
| 44 | d1d4m1_ |
|
not modelled |
7.8 |
57 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 45 | c2jg6A_ |
|
not modelled |
7.8 |
6 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna-3-methyladenine glycosidase;
PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
|
| 46 | c2lciA_ |
|
not modelled |
7.7 |
33 |
PDB header:de novo protein
Chain: A: PDB Molecule:protein or36;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or36 (casd3 target)
|
| 47 | d1jc4a_ |
|
not modelled |
7.6 |
12 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Methylmalonyl-CoA epimerase |
| 48 | c2rbbB_ |
|
not modelled |
7.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxalase/bleomycin resistance protein/dioxygenase;
PDBTitle: crystal structure of a glyoxalase/bleomycin resistance2 protein/dioxygenase family enzyme from burkholderia phytofirmans psjn
|
| 49 | d1pvc1_ |
|
not modelled |
7.6 |
25 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 50 | c2gcfA_ |
|
not modelled |
7.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:cation-transporting atpase pacs;
PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
|
| 51 | c3sviA_ |
|
not modelled |
7.4 |
36 |
PDB header:signaling protein
Chain: A: PDB Molecule:type iii effector hopab2;
PDBTitle: structure of the pto-binding domain of hoppmal generated by limited2 thermolysin digestion
|
| 52 | d1rhi1_ |
|
not modelled |
7.4 |
43 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 53 | c1sg3A_ |
|
not modelled |
7.4 |
56 |
PDB header:hydrolase
Chain: A: PDB Molecule:allantoicase;
PDBTitle: structure of allantoicase
|
| 54 | d1bjna_ |
|
not modelled |
7.3 |
9 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 55 | d2pc6a2 |
|
not modelled |
7.2 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 56 | c2kyzA_ |
|
not modelled |
7.2 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:heavy metal binding protein;
PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
|
| 57 | d2qifa1 |
|
not modelled |
7.2 |
24 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 58 | d2f1fa1 |
|
not modelled |
7.2 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 59 | c2k7wB_ |
|
not modelled |
7.1 |
19 |
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: bax activation is initiated at a novel interaction site
|
| 60 | c2rk0B_ |
|
not modelled |
7.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxalase/bleomycin resistance protein/dioxygenase domain;
PDBTitle: crystal structure of glyoxylase/bleomycin resistance2 protein/dioxygenase domain from frankia sp. ean1pec
|
| 61 | d1xqaa_ |
|
not modelled |
7.0 |
21 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Antibiotic resistance proteins |
| 62 | d1eah1_ |
|
not modelled |
7.0 |
19 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 63 | d1qqp1_ |
|
not modelled |
6.9 |
29 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 64 | c1ik9C_ |
|
not modelled |
6.8 |
50 |
PDB header:gene regulation/ligase
Chain: C: PDB Molecule:dna ligase iv;
PDBTitle: crystal structure of a xrcc4-dna ligase iv complex
|
| 65 | d1zhva2 |
|
not modelled |
6.8 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
| 66 | d1cov1_ |
|
not modelled |
6.8 |
57 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 67 | d2ajta2 |
|
not modelled |
6.7 |
10 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
| 68 | c1fi8E_ |
|
not modelled |
6.6 |
57 |
PDB header:hydrolase/hydrolase inhibitor
Chain: E: PDB Molecule:ecotin;
PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
|
| 69 | c3hgkE_ |
|
not modelled |
6.6 |
22 |
PDB header:transferase
Chain: E: PDB Molecule:effector protein hopab2;
PDBTitle: crystal structure of effect protein avrptob complexed with2 kinase pto
|
| 70 | c2r6uB_ |
|
not modelled |
6.6 |
45 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of gene product rha04853 from rhodococcus sp. rha1
|
| 71 | c3eyiB_ |
|
not modelled |
6.6 |
41 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:z-dna-binding protein 1;
PDBTitle: the crystal structure of the second z-dna binding domain of2 human dai (zbp1) in complex with z-dna
|
| 72 | c2jz7A_ |
|
not modelled |
6.5 |
28 |
PDB header:selenium-binding protein
Chain: A: PDB Molecule:selenium binding protein;
PDBTitle: solution nmr structure of selenium-binding protein from2 methanococcus vannielii
|
| 73 | d1zyea1 |
|
not modelled |
6.5 |
36 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 74 | c3q87B_ |
|
not modelled |
6.4 |
11 |
PDB header:transferase activator/transferase
Chain: B: PDB Molecule:n6 adenine specific dna methylase;
PDBTitle: structure of eukaryotic translation termination complex2 methyltransferase mtq2-trm112
|
| 75 | c3kolA_ |
|
not modelled |
6.4 |
64 |
PDB header:metal binding protein
Chain: A: PDB Molecule:glyoxalase/bleomycin resistance
PDBTitle: crystal structure of a glyoxalase/dioxygenase from nostoc2 punctiforme
|
| 76 | d2e7ja1 |
|
not modelled |
6.4 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
| 77 | d1aym1_ |
|
not modelled |
6.0 |
43 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 78 | c3oajA_ |
|
not modelled |
5.9 |
31 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative ring-cleaving dioxygenase mhqo;
PDBTitle: crystal structure of putative dioxygenase from bacillus subtilis2 subsp. subtilis str. 168
|
| 79 | c2nl9B_ |
|
not modelled |
5.9 |
19 |
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: crystal structure of the mcl-1:bim bh3 complex
|
| 80 | d1sqia2 |
|
not modelled |
5.9 |
15 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Extradiol dioxygenases |
| 81 | c3sk1C_ |
|
not modelled |
5.8 |
36 |
PDB header:griseoluteate-binding protein
Chain: C: PDB Molecule:ehpr;
PDBTitle: crystal structure of phenazine resistance protein ehpr from2 enterobacter agglomerans (erwinia herbicola, pantoea agglomerans)3 eh1087, apo form
|
| 82 | c2l69A_ |
|
not modelled |
5.8 |
27 |
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
|
| 83 | d1elua_ |
|
not modelled |
5.8 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 84 | d1tme1_ |
|
not modelled |
5.8 |
25 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 85 | c2cosA_ |
|
not modelled |
5.7 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase lats2;
PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
|
| 86 | d2ggpb1 |
|
not modelled |
5.5 |
10 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 87 | c1yg0A_ |
|
not modelled |
5.5 |
22 |
PDB header:metal transport
Chain: A: PDB Molecule:cop associated protein;
PDBTitle: solution structure of apo-copp from helicobacter pylori
|
| 88 | c3h25A_ |
|
not modelled |
5.5 |
18 |
PDB header:replication/dna
Chain: A: PDB Molecule:replication protein b;
PDBTitle: crystal structure of the catalytic domain of primase repb' in complex2 with initiator dna
|
| 89 | d1e2ya_ |
|
not modelled |
5.5 |
36 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 90 | d1kn0a_ |
|
not modelled |
5.4 |
23 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:The homologous-pairing domain of Rad52 recombinase |
| 91 | c3q1jA_ |
|
not modelled |
5.3 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
|
| 92 | c3rriB_ |
|
not modelled |
5.3 |
18 |
PDB header:metal binding protein
Chain: B: PDB Molecule:glyoxalase/bleomycin resistance protein/dioxygenase;
PDBTitle: crystal structure of glyoxalase/bleomycin resistance2 protein/dioxygenase from alicyclobacillus acidocaldarius
|
| 93 | d1t47a2 |
|
not modelled |
5.2 |
11 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Extradiol dioxygenases |
| 94 | d1k4na_ |
|
not modelled |
5.2 |
13 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Hypothetical protein YecM (EC4020) |
| 95 | c3e0rC_ |
|
not modelled |
5.2 |
11 |
PDB header:hydrolase
Chain: C: PDB Molecule:c3-degrading proteinase (cppa protein);
PDBTitle: crystal structure of cppa protein from streptococcus pneumoniae tigr4
|
| 96 | d1ug7a_ |
|
not modelled |
5.2 |
50 |
Fold:Four-helical up-and-down bundle
Superfamily:Domain from hypothetical 2610208m17rik protein
Family:Domain from hypothetical 2610208m17rik protein |
| 97 | d1tmf1_ |
|
not modelled |
5.1 |
15 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 98 | c2f1fA_ |
|
not modelled |
5.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
| 99 | d1kona_ |
|
not modelled |
5.0 |
20 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |