1 c1oy8A_
100.0
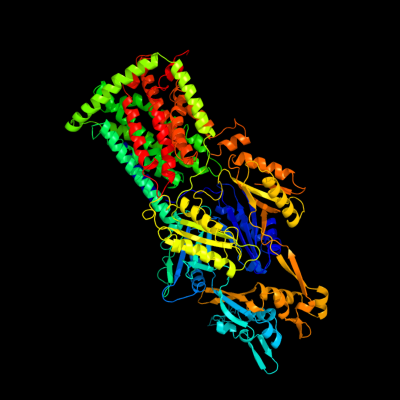
72
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
2 c3k07A_
100.0
21
PDB header: transport proteinChain: A: PDB Molecule: cation efflux system protein cusa;PDBTitle: crystal structure of cusa
3 c3aqpB_
100.0
17
PDB header: membrane proteinChain: B: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
4 d1iwga8
100.0
72
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain5 d1iwga7
100.0
73
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain6 d1iwga1
99.9
76
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains7 d1iwga2
99.7
39
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains8 d1iwga5
99.6
68
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsSuperfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsFamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains9 d1iwga6
99.2
56
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsSuperfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsFamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains10 d1iwga3
98.8
65
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains11 d1iwga4
98.2
75
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains12 c2ew9A_
96.4
16
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of apowln5-6
13 c2ropA_
95.9
15
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of domains 3 and 4 of human atp7b
14 c2rmlA_
94.7
17
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting p-type atpase copa;PDBTitle: solution structure of the n-terminal soluble domains of2 bacillus subtilis copa
15 d1s6ua_
91.8
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain16 d2aw0a_
91.6
22
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain17 d1cpza_
91.2
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain18 d2qifa1
91.1
17
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain19 d1q8la_
91.0
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain20 c3dxsX_
90.6
17
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
21 c2l3mA_
not modelled
90.5
17
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
22 d1p6ta1
not modelled
90.1
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain23 d1p6ta2
not modelled
90.0
17
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain24 d1kvja_
not modelled
89.8
22
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain25 c1yjrA_
not modelled
89.5
22
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
26 c2kkhA_
not modelled
89.3
13
PDB header: metal transportChain: A: PDB Molecule: putative heavy metal transporter;PDBTitle: structure of the zinc binding domain of the atpase hma4
27 c2ga7A_
not modelled
89.3
20
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
28 c1yg0A_
not modelled
89.2
9
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
29 c2ldiA_
not modelled
88.6
22
PDB header: hydrolaseChain: A: PDB Molecule: zinc-transporting atpase;PDBTitle: nmr solution structure of ziaan sub mutant
30 d1osda_
not modelled
88.5
17
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain31 c2ofhX_
not modelled
88.0
17
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
32 c2kt2A_
not modelled
87.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
33 d1afia_
not modelled
87.0
15
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain34 c1y3kA_
not modelled
84.0
20
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
35 d1qupa2
not modelled
83.1
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain36 c1jk9D_
not modelled
81.9
21
PDB header: oxidoreductaseChain: D: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: heterodimer between h48f-ysod1 and yccs
37 c2k2pA_
not modelled
81.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
38 d2ggpb1
not modelled
79.8
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain39 d1mwza_
not modelled
79.6
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain40 c2gcfA_
not modelled
78.0
18
PDB header: hydrolaseChain: A: PDB Molecule: cation-transporting atpase pacs;PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
41 d1sb6a_
not modelled
77.4
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain42 c3j09A_
not modelled
77.2
20
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
43 c2rogA_
not modelled
75.6
12
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
44 c1qupA_
not modelled
75.2
19
PDB header: chaperoneChain: A: PDB Molecule: superoxide dismutase 1 copper chaperone;PDBTitle: crystal structure of the copper chaperone for superoxide2 dismutase
45 c2crlA_
not modelled
74.7
16
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
46 c2yvxD_
not modelled
67.1
13
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
47 d1cc8a_
not modelled
66.7
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain48 c2aj1A_
not modelled
66.5
9
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
49 c1yj7A_
not modelled
62.1
16
PDB header: protein transportChain: A: PDB Molecule: escj;PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
50 c2kyzA_
not modelled
51.7
15
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
51 c1ciiA_
not modelled
40.3
17
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
52 d2vv5a2
not modelled
37.9
15
Fold: Ferredoxin-likeSuperfamily: Mechanosensitive channel protein MscS (YggB), C-terminal domainFamily: Mechanosensitive channel protein MscS (YggB), C-terminal domain53 d2cu6a1
not modelled
37.8
20
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like54 d1uwda_
not modelled
36.6
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like55 c3c6fD_
not modelled
29.1
8
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: yetf protein;PDBTitle: crystal structure of protein bsu07140 from bacillus subtilis
56 c3gdzA_
not modelled
28.1
14
PDB header: ligaseChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of arginyl-trna synthetase from klebsiella2 pneumoniae subsp. pneumoniae
57 c3fryB_
not modelled
25.0
15
PDB header: hydrolaseChain: B: PDB Molecule: probable copper-exporting p-type atpase a;PDBTitle: crystal structure of the copa c-terminal metal binding domain
58 d1jb0i_
not modelled
24.6
19
Fold: Single transmembrane helixSuperfamily: Subunit VIII of photosystem I reaction centre, PsaIFamily: Subunit VIII of photosystem I reaction centre, PsaI59 c3lnoA_
not modelled
24.2
18
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
60 d1f7ua3
not modelled
21.2
13
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domainFamily: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domain61 d1fe0a_
not modelled
20.6
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain62 c2bbjB_
not modelled
19.6
9
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
63 c2yy3B_
not modelled
17.9
9
PDB header: translationChain: B: PDB Molecule: elongation factor 1-beta;PDBTitle: crystal structure of translation elongation factor ef-1 beta from2 pyrococcus horikoshii
64 d1ek8a_
not modelled
16.5
27
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF65 c3iplB_
not modelled
16.1
12
PDB header: ligaseChain: B: PDB Molecule: 2-succinylbenzoate--coa ligase;PDBTitle: crystal structure of o-succinylbenzoic acid-coa ligase from2 staphylococcus aureus subsp. aureus mu50
66 d1wqga1
not modelled
15.8
19
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF67 d1a87a_
not modelled
15.7
15
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin68 c1a87A_
not modelled
15.7
15
PDB header: bacteriocinChain: A: PDB Molecule: colicin n;PDBTitle: colicin n
69 d1ydxa2
not modelled
14.6
19
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain70 c3g7sA_
not modelled
13.8
18
PDB header: ligaseChain: A: PDB Molecule: long-chain-fatty-acid--coa ligase (fadd-1);PDBTitle: crystal structure of a long-chain-fatty-acid-coa ligase2 (fadd1) from archaeoglobus fulgidus
71 d1dd5a_
not modelled
13.6
15
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF72 d1eh1a_
not modelled
13.5
23
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF73 d2phcb2
not modelled
13.5
16
Fold: DCoH-likeSuperfamily: PH0987 N-terminal domain-likeFamily: PH0987 N-terminal domain-like74 c2ki0A_
not modelled
13.2
13
PDB header: de novo proteinChain: A: PDB Molecule: ds119;PDBTitle: nmr structure of a de novo designed beta alpha beta
75 d1is1a_
not modelled
13.2
27
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF76 d2nwwa1
not modelled
13.0
10
Fold: Proton glutamate symport proteinSuperfamily: Proton glutamate symport proteinFamily: Proton glutamate symport protein77 d2ffma1
not modelled
12.9
9
Fold: Hypothetical protein SAV1430Superfamily: Hypothetical protein SAV1430Family: Hypothetical protein SAV143078 c1f7uA_
not modelled
12.6
13
PDB header: ligase/rnaChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of the arginyl-trna synthetase complexed with the2 trna(arg) and l-arg
79 d1kfta_
not modelled
12.4
18
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Excinuclease UvrC C-terminal domain80 c1kftA_
not modelled
12.4
18
PDB header: dna binding proteinChain: A: PDB Molecule: excinuclease abc subunit c;PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
81 d2b3ya2
not modelled
12.2
43
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain82 c3g74B_
not modelled
12.1
42
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function;PDBTitle: crystal structure of a functionally unknown protein from eubacterium2 ventriosum atcc 27560
83 d1x2ia1
not modelled
11.7
15
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like84 c2y9jt_
not modelled
11.6
14
PDB header: protein transportChain: T: PDB Molecule: protein prgh;PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
85 c2k1hA_
not modelled
11.5
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ser13;PDBTitle: solution nmr structure of ser13 from staphylococcus epidermidis.2 northeast structural genomics consortium target ser13
86 c2vxaL_
not modelled
11.3
21
PDB header: flavoproteinChain: L: PDB Molecule: dodecin;PDBTitle: h.halophila dodecin in complex with riboflavin
87 d1nwaa_
not modelled
11.2
23
Fold: Ferredoxin-likeSuperfamily: Peptide methionine sulfoxide reductaseFamily: Peptide methionine sulfoxide reductase88 c1nwaA_
not modelled
11.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: peptide methionine sulfoxide reductase msra;PDBTitle: structure of mycobacterium tuberculosis methionine2 sulfoxide reductase a in complex with protein-bound3 methionine
89 d2cfxa2
not modelled
11.2
7
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain90 c2hdiA_
not modelled
11.1
32
PDB header: protein transport,antimicrobial proteinChain: A: PDB Molecule: colicin i receptor;PDBTitle: crystal structure of the colicin i receptor cir from e.coli in complex2 with receptor binding domain of colicin ia.
91 d1ge9a_
not modelled
10.9
15
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Ribosome recycling factor, RRFFamily: Ribosome recycling factor, RRF92 d1yvwa1
not modelled
10.9
27
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: HisE-like (PRA-PH)93 c1yvwD_
not modelled
10.9
27
PDB header: hydrolaseChain: D: PDB Molecule: phosphoribosyl-atp pyrophosphatase;PDBTitle: crystal structure of phosphoribosyl-atp2 pyrophosphohydrolase from bacillus cereus. nesgc target3 bcr13.
94 d2ux9a1
not modelled
10.8
14
Fold: Dodecin subunit-likeSuperfamily: Dodecin-likeFamily: Dodecin-like95 d2cyya2
not modelled
10.7
4
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain96 c3oqtP_
not modelled
10.7
10
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
97 d2cg4a2
not modelled
10.3
9
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain98 c2k9pA_
not modelled
10.3
16
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
99 d2axti1
not modelled
10.3
12
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like