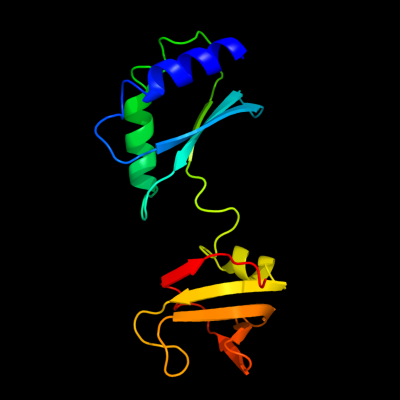
1 c1ib8A_
100.0
32
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: conserved protein sp14.3;PDBTitle: solution structure and function of a conserved protein2 sp14.3 encoded by an essential streptococcus pneumoniae3 gene
2 d1ib8a2
99.9
37
Fold: Alpha-lytic protease prodomain-likeSuperfamily: YhbC-like, N-terminal domainFamily: YhbC-like, N-terminal domain3 d1ib8a1
99.4
28
Fold: Sm-like foldSuperfamily: YhbC-like, C-terminal domainFamily: YhbC-like, C-terminal domain4 c1y96C_
88.7
11
PDB header: rna binding proteinChain: C: PDB Molecule: gem-associated protein 6;PDBTitle: crystal structure of the gemin6/gemin7 heterodimer from the2 human smn complex
5 c3cw15_
85.0
22
PDB header: splicingChain: 5: PDB Molecule: small nuclear ribonucleoprotein g;PDB Fragment: residues 1-215;
PDBTitle: crystal structure of human spliceosomal u1 snrnp
6 d1h641_
83.9
24
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP7 d1th7a1
83.0
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP8 d1i8fa_
80.8
18
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP9 d1m5q1_
79.4
37
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP10 d1mgqa_
77.4
25
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP11 c3bw1A_
77.1
29
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm3;PDBTitle: crystal structure of homomeric yeast lsm3 exhibiting novel octameric2 ring organisation
12 d1b34a_
76.3
32
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP13 c1b34A_
76.3
32
PDB header: rna binding proteinChain: A: PDB Molecule: protein (small nuclear ribonucleoprotein sm d1);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
14 d1jbma_
76.2
25
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP15 c3swnT_
75.8
21
PDB header: rna binding proteinChain: T: PDB Molecule: u6 snrna-associated sm-like protein lsm6;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
16 c3cw1Z_
74.9
25
PDB header: splicingChain: Z: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of human spliceosomal u1 snrnp
17 c1n9sH_
74.9
21
PDB header: translationChain: H: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of yeast smf in spacegroup p43212
18 c3swnA_
74.8
14
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
19 c2fwkB_
74.3
17
PDB header: dna binding proteinChain: B: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: crystal structure of cryptosporidium parvum u6 snrna-associated sm-2 like protein lsm5
20 d1ljoa_
74.2
17
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP21 d1d3bl_
not modelled
74.1
17
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP22 c3swnC_
not modelled
73.7
20
PDB header: rna binding proteinChain: C: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
23 d1n9ra_
not modelled
73.5
21
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP24 d1d3ba_
not modelled
72.6
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP25 d1n9sc_
not modelled
72.5
21
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP26 d1biaa2
not modelled
71.6
16
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)27 d1i4k1_
not modelled
71.3
20
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP28 c2ewnA_
not modelled
71.2
19
PDB header: ligase, transcriptionChain: A: PDB Molecule: bira bifunctional protein;PDBTitle: ecoli biotin repressor with co-repressor analog
29 c2e70A_
not modelled
70.9
13
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
30 d1d3bb_
not modelled
69.5
28
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP31 c2kvqG_
not modelled
68.4
30
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
32 c2jvvA_
not modelled
68.4
30
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
33 d1ycya1
not modelled
67.8
27
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: PF1955-like34 c3pgwQ_
not modelled
67.2
27
PDB header: splicing/dna/rnaChain: Q: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
35 d1nz9a_
not modelled
66.8
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain36 d2fwka1
not modelled
66.5
17
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP37 c3pgwB_
not modelled
64.3
26
PDB header: splicing/dna/rnaChain: B: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
38 c2ej9A_
not modelled
61.2
19
PDB header: ligaseChain: A: PDB Molecule: putative biotin ligase;PDBTitle: crystal structure of biotin protein ligase from2 methanococcus jannaschii
39 c3cw1A_
not modelled
58.8
27
PDB header: splicingChain: A: PDB Molecule: small nuclear ribonucleoprotein-associated proteins b andPDBTitle: crystal structure of human spliceosomal u1 snrnp
40 c2rm4A_
not modelled
56.7
25
PDB header: protein bindingChain: A: PDB Molecule: cg6311-pb;PDBTitle: solution structure of the lsm domain of dm edc3 (enhancer2 of decapping 3)
41 d1r5pa_
not modelled
52.9
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: KaiB-like42 c2cghB_
not modelled
52.5
18
PDB header: ligaseChain: B: PDB Molecule: biotin ligase;PDBTitle: crystal structure of biotin ligase from mycobacterium2 tuberculosis
43 c3cw1D_
not modelled
51.5
14
PDB header: splicingChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: crystal structure of human spliceosomal u1 snrnp
44 c2e6zA_
not modelled
47.4
30
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
45 d2pi2e1
not modelled
43.5
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB46 c2pqaB_
not modelled
42.0
26
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
47 d1qd1a1
not modelled
40.3
17
Fold: Ferredoxin-likeSuperfamily: Formiminotransferase domain of formiminotransferase-cyclodeaminase.Family: Formiminotransferase domain of formiminotransferase-cyclodeaminase.48 c2pt7G_
not modelled
39.2
17
PDB header: hydrolase/protein bindingChain: G: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of cag virb11 (hp0525) and an inhibitory protein2 (hp1451)
49 c2eayB_
not modelled
38.3
18
PDB header: ligaseChain: B: PDB Molecule: biotin [acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
50 d1nppa2
not modelled
36.4
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain51 d1kk8a1
not modelled
35.4
21
Fold: SH3-like barrelSuperfamily: Myosin S1 fragment, N-terminal domainFamily: Myosin S1 fragment, N-terminal domain52 d1h9aa1
not modelled
34.7
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain53 c2vc8A_
not modelled
34.5
24
PDB header: protein-bindingChain: A: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of the lsm domain of human edc3 (enhancer2 of decapping 3)
54 d1qkia1
not modelled
31.4
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain55 d1u2ma_
not modelled
31.3
13
Fold: OmpH-likeSuperfamily: OmpH-likeFamily: OmpH-like56 c2hbpA_
not modelled
29.1
12
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: solution structure of sla1 homology domain 1
57 c1tt9B_
not modelled
28.3
17
PDB header: transferase, lyaseChain: B: PDB Molecule: formimidoyltransferase-cyclodeaminasePDBTitle: structure of the bifunctional and golgi associated2 formiminotransferase cyclodeaminase octamer
58 d1b34b_
not modelled
26.9
35
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP59 c1b34B_
not modelled
26.9
35
PDB header: rna binding proteinChain: B: PDB Molecule: protein (small nuclear ribonucleoprotein sm d2);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
60 d1wina_
not modelled
25.4
15
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain61 d1sg5a1
not modelled
22.4
19
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: Rof-like62 c1gshA_
not modelled
22.3
16
PDB header: glutathione biosynthesis ligaseChain: A: PDB Molecule: glutathione biosynthetic ligase;PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
63 c1h9aA_
not modelled
22.1
18
PDB header: oxidoreductase (choh(d) - nad(p))Chain: A: PDB Molecule: glucose 6-phosphate 1-dehydrogenase;PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
64 d1ug7a_
not modelled
20.9
10
Fold: Four-helical up-and-down bundleSuperfamily: Domain from hypothetical 2610208m17rik proteinFamily: Domain from hypothetical 2610208m17rik protein65 c3cw1W_
not modelled
20.5
21
PDB header: splicingChain: W: PDB Molecule: small nuclear ribonucleoprotein e;PDBTitle: crystal structure of human spliceosomal u1 snrnp
66 c1qkiE_
not modelled
20.0
15
PDB header: oxidoreductaseChain: E: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase2 (variant canton r459l) complexed with structural nadp+
67 c3ic8D_
not modelled
20.0
20
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized gst-like proteinprotein;PDBTitle: the crystal structure of a gst-like protein from pseudomonas syringae2 to 2.4a
68 c2zy3A_
not modelled
18.2
13
PDB header: lyaseChain: A: PDB Molecule: l-aspartate beta-decarboxylase;PDBTitle: dodecameric l-aspartate beta-decarboxylase
69 d1t4za_
not modelled
17.5
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: KaiB-like70 d3bypa1
not modelled
17.2
11
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like71 d1kuta_
not modelled
17.1
25
Fold: SAICAR synthase-likeSuperfamily: SAICAR synthase-likeFamily: SAICAR synthase72 c2bhlB_
not modelled
16.8
15
PDB header: oxidoreductase (choh(d)-nadp)Chain: B: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase2 (deletion variant) complexed with glucose-6-phosphate
73 c3dexA_
not modelled
16.0
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sav_2001;PDBTitle: crystal structure of sav_2001 protein from streptomyces2 avermitilis, northeast structural genomics consortium3 target svr107.
74 c2iruA_
not modelled
16.0
8
PDB header: transferaseChain: A: PDB Molecule: putative dna ligase-like protein rv0938/mt0965;PDBTitle: crystal structure of the polymerase domain from mycobacterium2 tuberculosis ligase d
75 d1h8la1
not modelled
15.2
20
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain76 c1qd1A_
not modelled
15.1
19
PDB header: transferaseChain: A: PDB Molecule: formiminotransferase-cyclodeaminase;PDBTitle: the crystal structure of the formiminotransferase domain of2 formiminotransferase-cyclodeaminase.
77 c1zq1B_
not modelled
15.0
19
PDB header: lyaseChain: B: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit d;PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
78 c1bknA_
not modelled
14.9
28
PDB header: dna repairChain: A: PDB Molecule: mutl;PDBTitle: crystal structure of an n-terminal 40kd fragment of e. coli2 dna mismatch repair protein mutl
79 d2bu1a1
not modelled
14.8
26
Fold: RNA bacteriophage capsid proteinSuperfamily: RNA bacteriophage capsid proteinFamily: RNA bacteriophage capsid protein80 d1uwya1
not modelled
14.8
36
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain81 c2gqsA_
not modelled
14.5
21
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamidePDBTitle: saicar synthetase complexed with cair-mg2+ and adp
82 c3kreA_
not modelled
14.2
18
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamide synthase;PDBTitle: crystal structure of phosphoribosylaminoimidazole-succinocarboxamide2 synthase from ehrlichia chaffeensis at 1.8a resolution
83 c1m1gB_
not modelled
14.2
22
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
84 c2z02A_
not modelled
14.2
18
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamidePDBTitle: crystal structure of2 phosphoribosylaminoimidazolesuccinocarboxamide synthase3 wit atp from methanocaldococcus jannaschii
85 c2ywvB_
not modelled
13.7
4
PDB header: ligaseChain: B: PDB Molecule: phosphoribosylaminoimidazole succinocarboxamide synthetase;PDBTitle: crystal structure of saicar synthetase from geobacillus kaustophilus
86 c2faoB_
not modelled
13.5
14
PDB header: hydrolase/transferaseChain: B: PDB Molecule: probable atp-dependent dna ligase;PDBTitle: crystal structure of pseudomonas aeruginosa ligd polymerase2 domain
87 c3d0fA_
not modelled
13.4
9
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
88 c2h31A_
not modelled
13.3
18
PDB header: ligase, lyaseChain: A: PDB Molecule: multifunctional protein ade2;PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
89 c3nuaB_
not modelled
13.3
11
PDB header: ligaseChain: B: PDB Molecule: phosphoribosylaminoimidazole-succinocarboxamide synthase;PDBTitle: crystal structure of phosphoribosylaminoimidazole-succinocarboxamide2 synthase from clostridium perfringens
90 c2kz0A_
not modelled
13.1
9
PDB header: transcriptionChain: A: PDB Molecule: bola family protein;PDBTitle: solution structure of a bola protein (ech_0303) from ehrlichia2 chaffeensis. seattle structural genomics center for infectious3 disease target ehcha.10365.a
91 c3qiiA_
not modelled
12.7
18
PDB header: transcription regulatorChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
92 c3kbqA_
not modelled
12.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein ta0487;PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
93 c2p0gB_
not modelled
12.3
6
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: selenoprotein w-related protein;PDBTitle: crystal structure of selenoprotein w-related protein from2 vibrio cholerae. northeast structural genomics target vcr75
94 c2ojlB_
not modelled
12.0
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of q7waf1_borpa from bordetella parapertussis.2 northeast structural genomics target bpr68.
95 c3bfmA_
not modelled
11.9
20
PDB header: unknown functionChain: A: PDB Molecule: biotin protein ligase-like protein of unknown function;PDBTitle: crystal structure of a biotin protein ligase-like protein of unknown2 function (tm1040_0394) from silicibacter sp. tm1040 at 1.70 a3 resolution
96 c2ckkA_
not modelled
11.3
20
PDB header: nuclear proteinChain: A: PDB Molecule: kin17;PDBTitle: high resolution crystal structure of the human kin172 c-terminal domain containing a kow motif3 kin17.
97 d3bpda1
not modelled
11.3
8
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like98 d1utya_
not modelled
10.9
39
Fold: BTV NS2-like ssRNA-binding domainSuperfamily: BTV NS2-like ssRNA-binding domainFamily: BTV NS2-like ssRNA-binding domain99 c1utyA_
not modelled
10.9
39
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 2;PDBTitle: crystal structure of the rna binding domain of bluetongue2 virus non-structural protein 2(ns2)