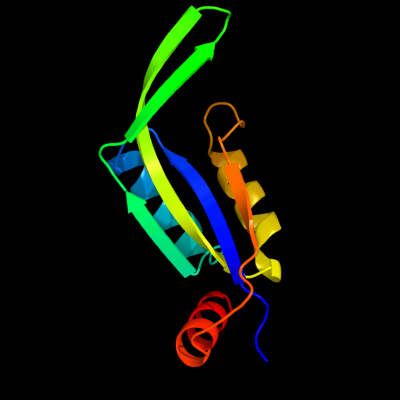
| 1 | d1naqa_
|
|
 |
100.0 |
100 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
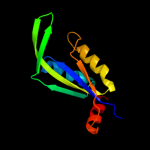
| 2 | c2zomC_
|
|
 |
100.0 |
41 |
PDB header:unknown function
Chain: C: PDB Molecule:protein cuta, chloroplast, putative, expressed;
PDBTitle: crystal structure of cuta1 from oryza sativa
|
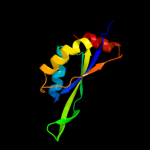
| 3 | c2zfhA_
|
|
 |
100.0 |
37 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cuta;
PDBTitle: crystal structure of putative cuta1 from homo sapiens at 2.05a2 resolution
|
| 4 | c1xk8A_
|
|
 |
100.0 |
37 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:divalent cation tolerant protein cuta;
PDBTitle: divalent cation tolerant protein cuta from homo sapiens2 o60888
|
| 5 | d2zfha1
|
|
 |
100.0 |
37 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 6 | c3ahpA_
|
|
 |
100.0 |
28 |
PDB header:electron transport
Chain: A: PDB Molecule:cuta1;
PDBTitle: crystal structure of stable protein, cuta1, from a psychrotrophic2 bacterium shewanella sp. sib1
|
| 7 | c2nuhA_
|
|
 |
100.0 |
36 |
PDB header:unknown function
Chain: A: PDB Molecule:periplasmic divalent cation tolerance protein;
PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
|
| 8 | d1ukua_
|
|
 |
100.0 |
37 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 9 | d1p1la_
|
|
 |
100.0 |
32 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 10 | d1kr4a_
|
|
 |
100.0 |
34 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 11 | d1osce_
|
|
 |
100.0 |
37 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 12 | d1nzaa_
|
|
 |
100.0 |
39 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 13 | d1vhfa_
|
|
 |
100.0 |
35 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 14 | c2gx8B_
|
|
 |
90.7 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:nif3-related protein;
PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
|
| 15 | d2gx8a1
|
|
 |
90.2 |
22 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
| 16 | c2nydB_
|
|
 |
72.8 |
20 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0135 protein sa1388;
PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
|
| 17 | d1v3fa_
|
|
 |
71.4 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 18 | d2csoa1
|
|
 |
62.7 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 19 | d1w4ma_
|
|
 |
36.8 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 20 | d1fsha_
|
|
 |
35.5 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 21 | d1uhwa_ |
|
not modelled |
35.2 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 22 | d1b4ba_ |
|
not modelled |
28.7 |
16 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 23 | c2pbiA_ |
|
not modelled |
28.6 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:regulator of g-protein signaling 9;
PDBTitle: the multifunctional nature of gbeta5/rgs9 revealed from its crystal2 structure
|
| 24 | c3cagF_ |
|
not modelled |
25.0 |
12 |
PDB header:dna binding protein
Chain: F: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the oligomerization domain hexamer of the2 arginine repressor protein from mycobacterium tuberculosis in complex3 with 9 arginines.
|
| 25 | d1gr0a1 |
|
not modelled |
22.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 26 | d1nkta4 |
|
not modelled |
22.8 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 27 | d1o7fa1 |
|
not modelled |
22.7 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
| 28 | c2ysrA_ |
|
not modelled |
21.4 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:dep domain-containing protein 1;
PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1
|
| 29 | c3tl6B_ |
|
not modelled |
18.5 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from entamoeba2 histolytica
|
| 30 | c1b4aA_ |
|
not modelled |
17.7 |
16 |
PDB header:repressor
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: structure of the arginine repressor from bacillus stearothermophilus
|
| 31 | d1iznb_ |
|
not modelled |
17.6 |
27 |
Fold:Subunits of heterodimeric actin filament capping protein Capz
Superfamily:Subunits of heterodimeric actin filament capping protein Capz
Family:Capz beta-1 subunit |
| 32 | c3lk2B_ |
|
not modelled |
17.5 |
27 |
PDB header:protein binding
Chain: B: PDB Molecule:f-actin-capping protein subunit beta isoforms 1 and 2;
PDBTitle: crystal structure of capz bound to the uncapping motif from carmil
|
| 33 | c3ml6D_ |
|
not modelled |
17.2 |
25 |
PDB header:protein transport
Chain: D: PDB Molecule:chimeric complex between protein dishevlled2 homolog dvl-2
PDBTitle: a complex between dishevlled2 and clathrin adaptor ap-2
|
| 34 | c3ereD_ |
|
not modelled |
17.1 |
12 |
PDB header:dna binding protein/dna
Chain: D: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the arginine repressor protein from mycobacterium2 tuberculosis in complex with the dna operator
|
| 35 | c1vytF_ |
|
not modelled |
13.8 |
33 |
PDB header:transport protein
Chain: F: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
|
| 36 | c1t0jC_ |
|
not modelled |
13.7 |
33 |
PDB header:signaling protein
Chain: C: PDB Molecule:voltage-dependent l-type calcium channel alpha-1c subunit;
PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
|
| 37 | d1u9ya1 |
|
not modelled |
12.8 |
12 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 38 | c3qmlC_ |
|
not modelled |
12.1 |
21 |
PDB header:chaperone/protein transport
Chain: C: PDB Molecule:nucleotide exchange factor sil1;
PDBTitle: the structural analysis of sil1-bip complex reveals the mechanism for2 sil1 to function as a novel nucleotide exchange factor
|
| 39 | c2v3sB_ |
|
not modelled |
12.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:serine/threonine-protein kinase osr1;
PDBTitle: structural insights into the recognition of substrates and2 activators by the osr1 kinase
|
| 40 | c2l23A_ |
|
not modelled |
11.8 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:mediator of rna polymerase ii transcription subunit 25;
PDBTitle: nmr structure of the acid (activator interacting domain) of the human2 mediator med25 protein
|
| 41 | d1i1ga2 |
|
not modelled |
11.6 |
18 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 42 | d2ixma1 |
|
not modelled |
10.9 |
18 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 43 | c2e1aD_ |
|
not modelled |
10.6 |
21 |
PDB header:transcription
Chain: D: PDB Molecule:75aa long hypothetical regulatory protein asnc;
PDBTitle: crystal structure of ffrp-dm1
|
| 44 | d2cfxa2 |
|
not modelled |
10.4 |
8 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 45 | d1vyua1 |
|
not modelled |
10.4 |
18 |
Fold:SH3-like barrel
Superfamily:SH3-domain
Family:SH3-domain |
| 46 | d1wjwa_ |
|
not modelled |
10.1 |
29 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 47 | c2zbcH_ |
|
not modelled |
9.7 |
24 |
PDB header:transcription
Chain: H: PDB Molecule:83aa long hypothetical transcriptional regulator asnc;
PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
|
| 48 | d1dkua1 |
|
not modelled |
9.7 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 49 | c1vytE_ |
|
not modelled |
9.3 |
33 |
PDB header:transport protein
Chain: E: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
|
| 50 | c3n5bB_ |
|
not modelled |
9.1 |
25 |
PDB header:transcription regulator
Chain: B: PDB Molecule:asr0485 protein;
PDBTitle: the complex of pii and pipx from anabaena
|
| 51 | c2g62A_ |
|
not modelled |
9.0 |
18 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:protein phosphatase 2a, regulatory subunit b' (pr 53);
PDBTitle: crystal structure of human ptpa
|
| 52 | c2xg8D_ |
|
not modelled |
8.9 |
17 |
PDB header:transcription
Chain: D: PDB Molecule:pipx;
PDBTitle: structural basis of gene regulation by protein pii: the2 crystal complex of pii and pipx from synechococcus3 elongatus pcc 7942
|
| 53 | c1ca0C_ |
|
not modelled |
8.0 |
25 |
PDB header:complex (serine protease/inhibitor)
Chain: C: PDB Molecule:bovine chymotrypsin;
PDBTitle: bovine chymotrypsin complexed to appi
|
| 54 | c2fugC_ |
|
not modelled |
7.7 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 55 | d1tf5a4 |
|
not modelled |
7.6 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 56 | c2djwF_ |
|
not modelled |
7.6 |
18 |
PDB header:unknown function
Chain: F: PDB Molecule:probable transcriptional regulator, asnc family;
PDBTitle: crystal structure of ttha0845 from thermus thermophilus hb8
|
| 57 | d1orfa_ |
|
not modelled |
7.6 |
13 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 58 | c2cy8A_ |
|
not modelled |
7.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:d-phenylglycine aminotransferase;
PDBTitle: crystal structure of d-phenylglycine aminotransferase (d-phgat) from2 pseudomonas strutzeri st-201
|
| 59 | d1xhna1 |
|
not modelled |
7.2 |
24 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 60 | d2p5ma1 |
|
not modelled |
7.1 |
14 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 61 | c3jx9B_ |
|
not modelled |
7.0 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
|
| 62 | d1fuja_ |
|
not modelled |
6.9 |
33 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 63 | d1pfxc_ |
|
not modelled |
6.7 |
17 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 64 | d1j6wa_ |
|
not modelled |
6.5 |
24 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 65 | c3h4wA_ |
|
not modelled |
6.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphatidylinositol-specific phospholipase c1;
PDBTitle: structure of a ca+2 dependent phosphatidylinositol-specific2 phospholipase c (pi-plc) enzyme from streptomyces antibioticus
|
| 66 | d1eufa_ |
|
not modelled |
6.2 |
40 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 67 | d2ac7a1 |
|
not modelled |
6.1 |
11 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 68 | d1m9ua_ |
|
not modelled |
6.1 |
33 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 69 | d1vjea_ |
|
not modelled |
6.0 |
41 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 70 | c2r0lA_ |
|
not modelled |
6.0 |
33 |
PDB header:hydrolase, immune system
Chain: A: PDB Molecule:hepatocyte growth factor activator;
PDBTitle: short form hgfa with inhibitory fab75
|
| 71 | d1fmca_ |
|
not modelled |
6.0 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | c1md7A_ |
|
not modelled |
5.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:c1r complement serine protease;
PDBTitle: monomeric structure of the zymogen of complement protease2 c1r
|
| 73 | c1x60A_ |
|
not modelled |
5.9 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:sporulation-specific n-acetylmuramoyl-l-alanine
PDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
|
| 74 | d1os8a_ |
|
not modelled |
5.8 |
17 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Prokaryotic proteases |
| 75 | d1xx9a_ |
|
not modelled |
5.8 |
20 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 76 | c1aksB_ |
|
not modelled |
5.6 |
20 |
PDB header:serine protease
Chain: B: PDB Molecule:alpha trypsin;
PDBTitle: crystal structure of the first active autolysate form of2 the porcine alpha trypsin
|
| 77 | d1xe0a_ |
|
not modelled |
5.5 |
32 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Nucleoplasmin-like core domain
Family:Nucleoplasmin-like core domain |
| 78 | d1qd1a2 |
|
not modelled |
5.4 |
8 |
Fold:Ferredoxin-like
Superfamily:Formiminotransferase domain of formiminotransferase-cyclodeaminase.
Family:Formiminotransferase domain of formiminotransferase-cyclodeaminase. |
| 79 | d1rfna_ |
|
not modelled |
5.3 |
17 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 80 | d2cyya2 |
|
not modelled |
5.2 |
22 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |