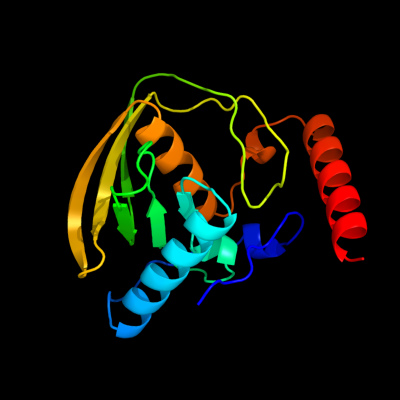
| 1 | c2w3tA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: chloro complex of the ni-form of e.coli deformylase
|
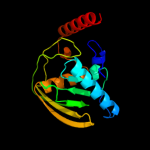
| 2 | d1ix1a_
|
|
 |
100.0 |
56 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
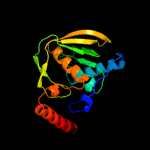
| 3 | d1xeoa1
|
|
 |
100.0 |
100 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 4 | c3qu1B_
|
|
 |
100.0 |
51 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:peptide deformylase 2;
PDBTitle: peptide deformylase from vibrio cholerae
|
| 5 | c3ocaB_
|
|
 |
100.0 |
42 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of peptide deformylase from ehrlichia chaffeensis
|
| 6 | c2ew7A_
|
|
 |
100.0 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of helicobacter pylori peptide deformylase
|
| 7 | c3cpmA_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase, chloroplast;
PDBTitle: plant peptide deformylase pdf1b crystal structure
|
| 8 | d1y6ha_
|
|
 |
100.0 |
35 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 9 | d1jyma_
|
|
 |
100.0 |
33 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 10 | c1ws1A_
|
|
 |
100.0 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase 1;
PDBTitle: structure analysis of peptide deformylase from bacillus2 cereus
|
| 11 | c3e3uA_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of mycobacterium tuberculosis peptide2 deformylase in complex with inhibitor
|
| 12 | d1rl4a_
|
|
 |
100.0 |
37 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 13 | c3g5pB_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptide deformylase, mitochondrial;
PDBTitle: structure and activity of human mitochondrial peptide deformylase, a2 novel cancer target
|
| 14 | d2defa_
|
|
 |
100.0 |
100 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 15 | d1lmea_
|
|
 |
100.0 |
39 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 16 | d1v3ya_
|
|
 |
100.0 |
33 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 17 | c3dldA_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of peptide deformylase, xoo1075, from2 xanthomonas oryzae pv. oryzae kacc10331
|
| 18 | d1lqya_
|
|
 |
100.0 |
30 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 19 | c3g6nA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of an efpdf complex with met-ala-ser
|
| 20 | c1zy1B_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptide deformylase, mitochondrial;
PDBTitle: x-ray structure of peptide deformylase from arabidopsis2 thaliana (atpdf1a) in complex with met-ala-ser
|
| 21 | d1lm4a_ |
|
not modelled |
100.0 |
29 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 22 | d1lm6a_ |
|
not modelled |
100.0 |
28 |
Fold:Peptide deformylase
Superfamily:Peptide deformylase
Family:Peptide deformylase |
| 23 | c3l87A_ |
|
not modelled |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: the crystal structure of smu.143c from streptococcus mutans ua159
|
| 24 | d1wmha_ |
|
not modelled |
38.5 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 25 | c2kvzA_ |
|
not modelled |
37.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ispe;
PDBTitle: structure of residues 161-235 of putative peptidoglycan binding2 protein lmo0835 from listeria monocytogenes: target lmr64b of the3 northeast structural genomics consortium
|
| 26 | d1pqsa_ |
|
not modelled |
34.5 |
20 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 27 | d1q1oa_ |
|
not modelled |
31.8 |
24 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 28 | d1quaa_ |
|
not modelled |
30.7 |
38 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 29 | d1ip9a_ |
|
not modelled |
26.9 |
26 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 30 | d1nd1a_ |
|
not modelled |
25.9 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 31 | d1wnia_ |
|
not modelled |
25.8 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 32 | d2bkfa1 |
|
not modelled |
25.2 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 33 | c1yp1A_ |
|
not modelled |
25.1 |
40 |
PDB header:hydrolase
Chain: A: PDB Molecule:fii;
PDBTitle: crystal structure of a non-hemorrhagic fibrin(ogen)olytic2 metalloproteinase from venom of agkistrodon acutus
|
| 34 | d1bswa_ |
|
not modelled |
25.0 |
50 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 35 | d1r55a_ |
|
not modelled |
24.1 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 36 | d4aiga_ |
|
not modelled |
23.7 |
40 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 37 | c3c37B_ |
|
not modelled |
21.6 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase, m48 family;
PDBTitle: x-ray structure of the putative zn-dependent peptidase q74d82 at the2 resolution 1.7 a. northeast structural genomics consortium target3 gsr143a
|
| 38 | d1atla_ |
|
not modelled |
21.4 |
31 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 39 | c3k7nA_ |
|
not modelled |
21.2 |
47 |
PDB header:hydrolase
Chain: A: PDB Molecule:k-like;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 40 | c2kt7A_ |
|
not modelled |
20.5 |
28 |
PDB header:cell adhesion, membrane protein
Chain: A: PDB Molecule:putative peptidoglycan bound protein (lpxtg
PDBTitle: solution nmr structure of mucin-binding domain of protein2 lmo0835 from listeria monocytogenes, northeast structural3 genomics consortium target lmr64a
|
| 41 | d1kufa_ |
|
not modelled |
20.5 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 42 | c3k7lA_ |
|
not modelled |
19.5 |
47 |
PDB header:hydrolase
Chain: A: PDB Molecule:atragin;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 43 | c2dw1B_ |
|
not modelled |
14.3 |
40 |
PDB header:apoptosis, toxin
Chain: B: PDB Molecule:catrocollastatin;
PDBTitle: crystal structure of vap2 from crotalus atrox venom (form 2-2 crystal)
|
| 44 | c3bmbB_ |
|
not modelled |
14.2 |
15 |
PDB header:rna binding protein
Chain: B: PDB Molecule:regulator of nucleoside diphosphate kinase;
PDBTitle: crystal structure of a new rna polymerase interacting2 protein
|
| 45 | c2erpA_ |
|
not modelled |
13.8 |
53 |
PDB header:toxin
Chain: A: PDB Molecule:vascular apoptosis-inducing protein 1;
PDBTitle: crystal structure of vascular apoptosis-inducing protein-1(inhibitor-2 bound form)
|
| 46 | d1c7ka_ |
|
not modelled |
12.9 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Zinc protease |
| 47 | c2e3xA_ |
|
not modelled |
12.7 |
33 |
PDB header:hydrolase, blood clotting, toxin
Chain: A: PDB Molecule:coagulation factor x-activating enzyme heavy chain;
PDBTitle: crystal structure of russell's viper venom metalloproteinase
|
| 48 | c3b8zB_ |
|
not modelled |
12.7 |
46 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein adamts-5;
PDBTitle: high resolution crystal structure of the catalytic domain2 of adamts-5 (aggrecanase-2)
|
| 49 | c1z5sD_ |
|
not modelled |
12.6 |
33 |
PDB header:ligase
Chain: D: PDB Molecule:ran-binding protein 2;
PDBTitle: crystal structure of a complex between ubc9, sumo-1,2 rangap1 and nup358/ranbp2
|
| 50 | d1ydla1 |
|
not modelled |
12.4 |
24 |
Fold:TFB5-like
Superfamily:TFB5-like
Family:TFB5-like |
| 51 | d1wj6a_ |
|
not modelled |
12.1 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 52 | d1ytqa1 |
|
not modelled |
12.0 |
24 |
Fold:gamma-Crystallin-like
Superfamily:gamma-Crystallin-like
Family:Crystallins/Ca-binding development proteins |
| 53 | d1cxva_ |
|
not modelled |
11.9 |
43 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 54 | d1a8ya3 |
|
not modelled |
11.3 |
20 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Calsequestrin |
| 55 | c2jz7A_ |
|
not modelled |
11.1 |
22 |
PDB header:selenium-binding protein
Chain: A: PDB Molecule:selenium binding protein;
PDBTitle: solution nmr structure of selenium-binding protein from2 methanococcus vannielii
|
| 56 | c2rjpC_ |
|
not modelled |
10.5 |
23 |
PDB header:hydrolase
Chain: C: PDB Molecule:adamts-4;
PDBTitle: crystal structure of adamts4 with inhibitor bound
|
| 57 | c3g5cA_ |
|
not modelled |
10.0 |
25 |
PDB header:membrane protein
Chain: A: PDB Molecule:adam 22;
PDBTitle: structural and biochemical studies on the ectodomain of human adam22
|
| 58 | d1xuca1 |
|
not modelled |
9.6 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 59 | d1v8ca2 |
|
not modelled |
9.3 |
54 |
Fold:TBP-like
Superfamily:MoaD-related protein, C-terminal domain
Family:MoaD-related protein, C-terminal domain |
| 60 | d1qiba_ |
|
not modelled |
9.0 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 61 | c1zv8B_ |
|
not modelled |
9.0 |
56 |
PDB header:viral protein
Chain: B: PDB Molecule:e2 glycoprotein;
PDBTitle: a structure-based mechanism of sars virus membrane fusion
|
| 62 | d1hv5a_ |
|
not modelled |
9.0 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 63 | d1i76a_ |
|
not modelled |
8.9 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 64 | d1rr8c1 |
|
not modelled |
8.7 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 65 | d1hova_ |
|
not modelled |
8.5 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 66 | d1hy7a_ |
|
not modelled |
8.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 67 | c2i47A_ |
|
not modelled |
8.2 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:adam 17;
PDBTitle: crystal structure of catalytic domain of tace with inhibitor
|
| 68 | c2xs4A_ |
|
not modelled |
8.1 |
45 |
PDB header:hydrolase
Chain: A: PDB Molecule:karilysin protease;
PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
|
| 69 | c2h7fX_ |
|
not modelled |
8.0 |
17 |
PDB header:isomerase/dna
Chain: X: PDB Molecule:dna topoisomerase 1;
PDBTitle: structure of variola topoisomerase covalently bound to dna
|
| 70 | c3nppA_ |
|
not modelled |
7.9 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pfam duf1093 family protein;
PDBTitle: crystal structure of a pfam duf1093 family protein (bsu39620) from2 bacillus subtilis at 2.15 a resolution
|
| 71 | d2k5wa1 |
|
not modelled |
7.9 |
22 |
Fold:OB-fold
Superfamily:BC4932-like
Family:BC4932-like |
| 72 | d1cb8a2 |
|
not modelled |
7.6 |
17 |
Fold:Hyaluronate lyase-like, C-terminal domain
Superfamily:Hyaluronate lyase-like, C-terminal domain
Family:Hyaluronate lyase-like, C-terminal domain |
| 73 | c2wd6B_ |
|
not modelled |
7.4 |
29 |
PDB header:cell adhesion
Chain: B: PDB Molecule:agglutinin receptor;
PDBTitle: crystal structure of the variable domain of the2 streptococcus gordonii surface protein sspb
|
| 74 | d1fbla2 |
|
not modelled |
7.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 75 | d1rm8a_ |
|
not modelled |
7.3 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 76 | c2ktrA_ |
|
not modelled |
7.2 |
13 |
PDB header:signaling protein, transport protein
Chain: A: PDB Molecule:sequestosome-1;
PDBTitle: nmr structure of p62 pb1 dimer determined based on pcs
|
| 77 | d2i47a1 |
|
not modelled |
7.1 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TNF-alpha converting enzyme, TACE, catalytic domain |
| 78 | d1y6kr2 |
|
not modelled |
6.9 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 79 | d1k4ta2 |
|
not modelled |
6.8 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 80 | d2fkia1 |
|
not modelled |
6.7 |
25 |
Fold:Secretion chaperone-like
Superfamily:YjbR-like
Family:YjbR-like |
| 81 | c2x7mA_ |
|
not modelled |
6.4 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:archaemetzincin;
PDBTitle: crystal structure of archaemetzincin (amza) from methanopyrus2 kandleri at 1.5 a resolution
|
| 82 | d1jmma_ |
|
not modelled |
6.3 |
38 |
Fold:Supersandwich
Superfamily:V-region of surface antigen I/II (SA I/II, PAC)
Family:V-region of surface antigen I/II (SA I/II, PAC) |
| 83 | c3uinD_ |
|
not modelled |
6.1 |
33 |
PDB header:ligase/isomerase/protein binding
Chain: D: PDB Molecule:e3 sumo-protein ligase ranbp2;
PDBTitle: complex between human rangap1-sumo2, ubc9 and the ir1 domain from2 ranbp2
|
| 84 | d2axoa1 |
|
not modelled |
5.8 |
57 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Atu2684-like |
| 85 | d2k5qa1 |
|
not modelled |
5.8 |
44 |
Fold:OB-fold
Superfamily:BC4932-like
Family:BC4932-like |
| 86 | d1bqqm_ |
|
not modelled |
5.7 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 87 | d1y93a1 |
|
not modelled |
5.6 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 88 | c2axoA_ |
|
not modelled |
5.5 |
57 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein atu2684;
PDBTitle: x-ray crystal structure of protein agr_c_4864 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr35.
|
| 89 | c1ytqA_ |
|
not modelled |
5.5 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:beta crystallin b2;
PDBTitle: structure of native human beta b2 crystallin
|
| 90 | d2ovxa1 |
|
not modelled |
5.4 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 91 | d1mmqa_ |
|
not modelled |
5.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |