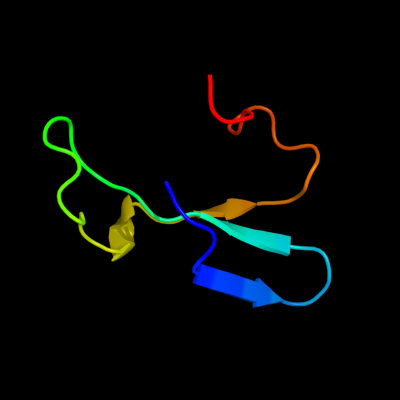
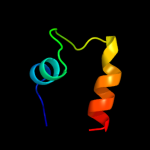
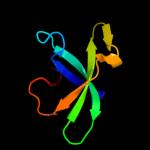
1 d1oz2a2
26.3
30
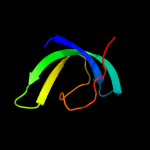
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat2 c2wzoA_
23.5
36
PDB header: cell cycleChain: A: PDB Molecule: transforming growth factor beta regulator 1;PDBTitle: the structure of the fyr domain
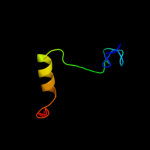
3 d1pkua1
23.4
15
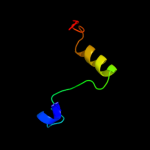
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK4 c2vytA_
16.5
33
PDB header: transcriptionChain: A: PDB Molecule: sex comb on midleg-like protein 2;PDBTitle: the mbt repeats of human scml2 bind to peptides containing2 mono methylated lysine.
5 c2r58A_
13.9
33
PDB header: transcriptionChain: A: PDB Molecule: polycomb protein scm;PDBTitle: crystal structure of the two mbt repeats from sex-comb on midleg (scm)2 in complex with di-methyl lysine
6 d1j6ra_
13.1
21
Fold: Methionine synthase activation domain-likeSuperfamily: Methionine synthase activation domain-likeFamily: Hypothetical protein TM02697 d1oi1a2
12.9
33
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat8 d1oi1a1
12.8
33
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat9 c2fkkA_
12.6
50
PDB header: viral proteinChain: A: PDB Molecule: baseplate structural protein gp10;PDBTitle: crystal structure of the c-terminal domain of the bacteriophage t42 gene product 10
10 c2eqmA_
12.1
19
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
11 d1wjra_
12.0
33
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat12 d1oz2a1
12.0
33
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat13 d1wjsa_
10.6
42
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat14 c2fl8N_
9.7
50
PDB header: virus/viral proteinChain: N: PDB Molecule: baseplate structural protein gp10;PDBTitle: fitting of the gp10 trimer structure into the cryoem map of the2 bacteriophage t4 baseplate in the hexagonal conformation.
15 d3bbba1
9.4
14
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK16 d1wjqa_
9.3
50
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat17 c3mpdA_
9.2
10
PDB header: transferaseChain: A: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: crystal structure of nucleoside diphosphate kinase from2 encephalitozoon cuniculi, cubic form, apo
18 d1u8wa_
8.9
10
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK19 d1oz2a3
8.5
20
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: MBT repeat20 c3feoB_
8.3
25
PDB header: metal binding proteinChain: B: PDB Molecule: mbt domain-containing protein 1;PDBTitle: the crystal structure of mbtd1
21 c3orqA_
not modelled
7.8
15
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
22 c3ceyA_
not modelled
7.7
42
PDB header: transcription regulatorChain: A: PDB Molecule: lethal(3)malignant brain tumor-like 2 protein;PDBTitle: crystal structure of l3mbtl2
23 c2ym9C_
not modelled
7.3
29
PDB header: cell invasionChain: C: PDB Molecule: cell invasion protein sipd;PDBTitle: sipd from salmonella typhimurium
24 d1m9fd_
not modelled
7.3
35
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain25 d1q8ha_
not modelled
6.9
71
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain26 c1q8hA_
not modelled
6.9
71
PDB header: metal binding proteinChain: A: PDB Molecule: osteocalcin;PDBTitle: crystal structure of porcine osteocalcin
27 c3kztB_
not modelled
6.7
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (np_812423.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
28 c3ngtJ_
not modelled
6.5
11
PDB header: transferaseChain: J: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: structure of leishmania ndkb complexed with amp.
29 c1oz3C_
not modelled
6.5
60
PDB header: transcriptionChain: C: PDB Molecule: lethal(3)malignant brain tumor-like protein;PDBTitle: crystal structure of 3-mbt repeats of lethal (3) malignant brain tumor2 (native-i) at 1.85 angstrom
30 d2jaaa1
not modelled
6.4
18
Fold: IpaD-likeSuperfamily: IpaD-likeFamily: IpaD-like31 c3ut1A_
not modelled
6.4
50
PDB header: transcriptionChain: A: PDB Molecule: lethal(3)malignant brain tumor-like protein 3;PDBTitle: crystal structure of the 3-mbt repeat domain of l3mbtl3
32 d1q3ma_
not modelled
6.3
71
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain33 d2o8ra3
not modelled
6.2
25
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain34 c3bp1A_
not modelled
6.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph-dependent 7-cyano-7-deazaguanine reductase;PDBTitle: crystal structure of putative 7-cyano-7-deazaguanine2 reductase quef from vibrio cholerae o1 biovar eltor
35 d2py5a1
not modelled
6.2
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease36 d2pxrc1
not modelled
6.2
35
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain37 d1kp0a1
not modelled
6.0
33
Fold: Ribonuclease H-like motifSuperfamily: Creatinase/prolidase N-terminal domainFamily: Creatinase/prolidase N-terminal domain38 d1s57a_
not modelled
5.8
13
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK39 c1j1eB_
not modelled
5.8
14
PDB header: contractile proteinChain: B: PDB Molecule: troponin t;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
40 c3zvmA_
not modelled
5.6
18
PDB header: hydrolase/transferase/dnaChain: A: PDB Molecule: bifunctional polynucleotide phosphatase/kinase;PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
41 c3nzzA_
not modelled
5.5
29
PDB header: cell invasionChain: A: PDB Molecule: cell invasion protein sipd;PDBTitle: crystal structure of the salmonella type iii secretion system tip2 protein sipd
42 c2kn8A_
not modelled
5.5
43
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: dna cleavage and packaging protein large subunit, ul89;PDBTitle: nmr structure of the c-terminal domain of pul89
43 c3r9lA_
not modelled
5.5
11
PDB header: transferaseChain: A: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: crystal structure of nucleoside diphosphate kinase from giardia2 lamblia featuring a disordered dinucleotide binding site
44 d1iowa1
not modelled
5.5
20
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain45 c1j1eC_
not modelled
5.2
47
PDB header: contractile proteinChain: C: PDB Molecule: troponin i;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
46 c2izpB_
not modelled
5.1
29
PDB header: toxinChain: B: PDB Molecule: putative membrane antigen;PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.