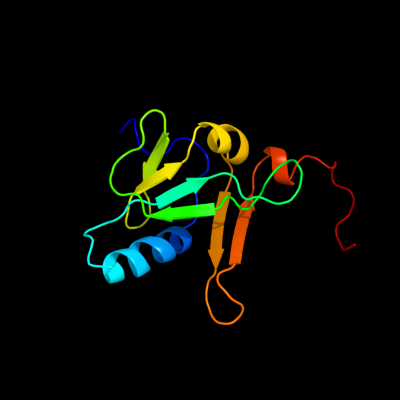
| 1 | c2apnA_
|
|
 |
100.0 |
82 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein hi1723;
PDBTitle: hi1723 solution structure
|
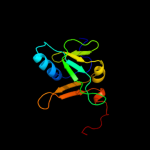
| 2 | c2d2aA_
|
|
 |
100.0 |
29 |
PDB header:metal transport
Chain: A: PDB Molecule:sufa protein;
PDBTitle: crystal structure of escherichia coli sufa involved in2 biosynthesis of iron-sulfur clusters
|
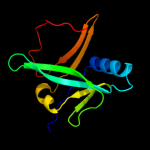
| 3 | c1x0gA_
|
|
 |
100.0 |
32 |
PDB header:metal binding protein
Chain: A: PDB Molecule:isca;
PDBTitle: crystal structure of isca with the [2fe-2s] cluster
|
| 4 | d1s98a_
|
|
 |
100.0 |
33 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 5 | c2k4zA_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsrr;
PDBTitle: solution nmr structure of allochromatium vinosum dsrr:2 northeast structural genomics consortium target op5
|
| 6 | d1nwba_
|
|
 |
100.0 |
38 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 7 | d2p2ea1
|
|
 |
98.7 |
20 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 8 | c2qgoA_
|
|
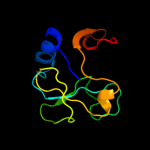 |
98.6 |
23 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative fe-s biosynthesis protein;
PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
|
| 9 | d1vsra_
|
|
 |
23.3 |
7 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Very short patch repair (VSR) endonuclease |
| 10 | d1sp2a_
|
|
 |
19.1 |
44 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 11 | c2kvfA_
|
|
 |
17.5 |
45 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger and btb domain-containing protein 32;
PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
|
| 12 | d1zfda_
|
|
 |
16.9 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 13 | d1a1ia1
|
|
 |
15.4 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 14 | d1aaya1
|
|
 |
14.9 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 15 | d1ubdc3
|
|
 |
14.5 |
71 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 16 | d1a1ha1
|
|
 |
13.9 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 17 | c2nq2C_
|
|
 |
13.1 |
20 |
PDB header:metal transport
Chain: C: PDB Molecule:hypothetical abc transporter atp-binding protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
|
| 18 | d1a1ga1
|
|
 |
13.1 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 19 | d1tf3a2
|
|
 |
13.1 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 20 | d1f2ig1
|
|
 |
12.7 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 21 | d1u86a1 |
|
not modelled |
12.6 |
36 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 22 | d2a6sa1 |
|
not modelled |
12.6 |
5 |
Fold:RelE-like
Superfamily:RelE-like
Family:YoeB/Txe-like |
| 23 | c1bcrA_ |
|
not modelled |
12.3 |
28 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
|
| 24 | d2ns1b1 |
|
not modelled |
12.1 |
10 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 25 | d1wmia1 |
|
not modelled |
12.0 |
11 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
| 26 | c3kxeD_ |
|
not modelled |
11.9 |
20 |
PDB header:protein binding
Chain: D: PDB Molecule:antitoxin protein pard-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
| 27 | d2yt9a1 |
|
not modelled |
11.2 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 28 | d1x6ea2 |
|
not modelled |
10.1 |
67 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 29 | d2glia3 |
|
not modelled |
10.1 |
36 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 30 | c3g5oC_ |
|
not modelled |
10.0 |
24 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
| 31 | c1znmA_ |
|
not modelled |
9.6 |
50 |
PDB header:zinc finger
Chain: A: PDB Molecule:yy1;
PDBTitle: a zinc finger with an artificial beta-turn, original2 sequence taken from the third zinc finger domain of the3 human transcriptional repressor protein yy1 (ying and yang4 1, a delta transcription factor), nmr, 34 structures
|
| 32 | d2dlka2 |
|
not modelled |
9.3 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 33 | d1ncsa_ |
|
not modelled |
9.3 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 34 | c2ab3A_ |
|
not modelled |
9.1 |
67 |
PDB header:rna binding protein
Chain: A: PDB Molecule:znf29;
PDBTitle: solution structures and characterization of hiv rre iib rna2 targeting zinc finger proteins
|
| 35 | c2kdxA_ |
|
not modelled |
8.7 |
13 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
| 36 | d1jj7a_ |
|
not modelled |
8.5 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 37 | d1wgea1 |
|
not modelled |
8.3 |
50 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 38 | c2kdnA_ |
|
not modelled |
8.2 |
60 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein pfe0790c;
PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
|
| 39 | d1m0sa2 |
|
not modelled |
8.1 |
23 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 40 | d1v9ja_ |
|
not modelled |
7.7 |
20 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:BolA-like
Family:BolA-like |
| 41 | d1lk5a2 |
|
not modelled |
7.6 |
16 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 42 | c1va1A_ |
|
not modelled |
7.6 |
43 |
PDB header:transcription
Chain: A: PDB Molecule:transcription factor sp1;
PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 1)
|
| 43 | c1zc1A_ |
|
not modelled |
7.4 |
21 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
| 44 | d2cota1 |
|
not modelled |
7.3 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 45 | d1ji0a_ |
|
not modelled |
7.2 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 46 | c2kheA_ |
|
not modelled |
7.2 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
|
| 47 | c2jn4A_ |
|
not modelled |
7.1 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein fixu, nift;
PDBTitle: solution nmr structure of protein rp4601 from2 rhodopseudomonas palustris. northeast structural genomics3 consortium target rpt2; ontario center for structural4 proteomics target rp4601.
|
| 48 | d2jn4a1 |
|
not modelled |
7.1 |
22 |
Fold:NifT/FixU barrel-like
Superfamily:NifT/FixU-like
Family:NifT/FixU |
| 49 | d2hq2a1 |
|
not modelled |
7.1 |
20 |
Fold:Heme iron utilization protein-like
Superfamily:Heme iron utilization protein-like
Family:HemS/ChuS-like |
| 50 | c3a44D_ |
|
not modelled |
7.1 |
20 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
| 51 | c2wh44_ |
|
not modelled |
6.9 |
67 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome
|
| 52 | c2yujA_ |
|
not modelled |
6.5 |
28 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
|
| 53 | d1ywsa1 |
|
not modelled |
6.4 |
56 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 54 | c3f1f4_ |
|
not modelled |
6.2 |
67 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: crystal structure of a translation termination complex2 formed with release factor rf2. this file contains the 50s3 subunit of one 70s ribosome. the entire crystal structure4 contains two 70s ribosomes as described in remark 400.
|
| 55 | d2j0pa1 |
|
not modelled |
6.2 |
17 |
Fold:Heme iron utilization protein-like
Superfamily:Heme iron utilization protein-like
Family:HemS/ChuS-like |
| 56 | d2j7ja1 |
|
not modelled |
6.2 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 57 | c1gxsC_ |
|
not modelled |
6.2 |
23 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 58 | d2a2ua_ |
|
not modelled |
6.2 |
9 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 59 | c3hlzA_ |
|
not modelled |
6.2 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bt_1490;
PDBTitle: crystal structure of bt_1490 (np_810393.1) from bacteroides2 thetaiotaomicron vpi-5482 at 1.50 a resolution
|
| 60 | c2jr7A_ |
|
not modelled |
6.1 |
67 |
PDB header:metal binding protein
Chain: A: PDB Molecule:dph3 homolog;
PDBTitle: solution structure of human desr1
|
| 61 | c3kxeB_ |
|
not modelled |
6.1 |
12 |
PDB header:protein binding
Chain: B: PDB Molecule:toxin protein pare-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
| 62 | c2cbzA_ |
|
not modelled |
5.9 |
26 |
PDB header:transport
Chain: A: PDB Molecule:multidrug resistance-associated protein 1;
PDBTitle: structure of the human multidrug resistance protein 12 nucleotide binding domain 1
|
| 63 | d1a1ia3 |
|
not modelled |
5.9 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 64 | d2cota2 |
|
not modelled |
5.9 |
83 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 65 | d3b60a1 |
|
not modelled |
5.9 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 66 | d1adta2 |
|
not modelled |
5.7 |
40 |
Fold:Zn-binding domains of ADDBP
Superfamily:Zn-binding domains of ADDBP
Family:Zn-binding domains of ADDBP |
| 67 | d1ubdc2 |
|
not modelled |
5.7 |
60 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 68 | d2j0141 |
|
not modelled |
5.6 |
40 |
Fold:L28p-like
Superfamily:L28p-like
Family:Ribosomal protein L31p |
| 69 | c2j034_ |
|
not modelled |
5.6 |
40 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
|
| 70 | d1ewfa1 |
|
not modelled |
5.6 |
0 |
Fold:Aha1/BPI domain-like
Superfamily:Bactericidal permeability-increasing protein, BPI
Family:Bactericidal permeability-increasing protein, BPI |
| 71 | c3gfoA_ |
|
not modelled |
5.6 |
18 |
PDB header:atp binding protein
Chain: A: PDB Molecule:cobalt import atp-binding protein cbio 1;
PDBTitle: structure of cbio1 from clostridium perfringens: part of2 the abc transporter complex cbionq.
|
| 72 | d1u85a1 |
|
not modelled |
5.6 |
67 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 73 | c3a0rA_ |
|
not modelled |
5.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
| 74 | c3c3jA_ |
|
not modelled |
5.5 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
|
| 75 | d1llmc2 |
|
not modelled |
5.5 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 76 | c2entA_ |
|
not modelled |
5.4 |
63 |
PDB header:transcription
Chain: A: PDB Molecule:krueppel-like factor 15;
PDBTitle: solution structure of the second c2h2-type zinc finger2 domain from human krueppel-like factor 15
|
| 77 | c2k6lA_ |
|
not modelled |
5.2 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
|
| 78 | c3t7lA_ |
|
not modelled |
5.2 |
21 |
PDB header:transport protein
Chain: A: PDB Molecule:zinc finger fyve domain-containing protein 16;
PDBTitle: crystal structure of the fyve domain of endofin (zfyve16) at 1.1a2 resolution
|
| 79 | d2h85a2 |
|
not modelled |
5.2 |
43 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 80 | c2l72A_ |
|
not modelled |
5.0 |
6 |
PDB header:protein binding
Chain: A: PDB Molecule:actin depolymerizing factor, putative;
PDBTitle: solution structure and dynamics of adf from toxoplasma gondii (tgadf)
|
| 81 | c1areA_ |
|
not modelled |
5.0 |
33 |
PDB header:transcription regulation
Chain: A: PDB Molecule:yeast transcription factor adr1;
PDBTitle: structures of dna-binding mutant zinc finger domains:2 implications for dna binding
|