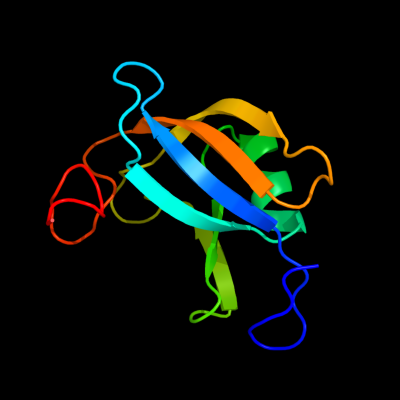
| 1 | d1zpsa1
|
|
 |
100.0 |
37 |
Fold:HisI-like
Superfamily:HisI-like
Family:HisI-like |
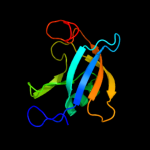
| 2 | d2a7wa1
|
|
 |
100.0 |
36 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
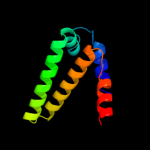
| 3 | c2a7wF_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: F: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
|
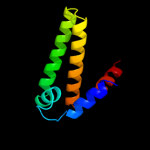
| 4 | d1yvwa1
|
|
 |
100.0 |
32 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 5 | c1yvwD_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: D: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp2 pyrophosphohydrolase from bacillus cereus. nesgc target3 bcr13.
|
| 6 | d1y6xa1
|
|
 |
100.0 |
30 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 7 | d1yxba1
|
|
 |
99.9 |
33 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 8 | c3crcB_
|
|
 |
98.8 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein mazg;
PDBTitle: crystal structure of escherichia coli mazg, the regulator2 of nutritional stress response
|
| 9 | c2yxhB_
|
|
 |
98.8 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:mazg-related protein;
PDBTitle: crystal structure of mazg-related protein from thermotoga maritima
|
| 10 | d1vmga_
|
|
 |
98.5 |
19 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 11 | d2a3qa1
|
|
 |
98.3 |
23 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 12 | c3obcB_
|
|
 |
98.3 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:pyrophosphatase;
PDBTitle: crystal structure of a pyrophosphatase (af1178) from archaeoglobus2 fulgidus at 1.80 a resolution
|
| 13 | d2gtad1
|
|
 |
98.3 |
15 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 14 | c2q4pA_
|
|
 |
98.3 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein rs21-c6;
PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898
|
| 15 | d2gtaa1
|
|
 |
98.2 |
16 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 16 | c2q9lA_
|
|
 |
98.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of imazg from vibrio dat 722: ctag-imazg (p43212)
|
| 17 | d2oiea1
|
|
 |
98.0 |
22 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 18 | c2yf3F_
|
|
 |
96.3 |
25 |
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from2 deinococcus radiodurans, complex with manganese
|
| 19 | c2rfpA_
|
|
 |
84.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative ntp pyrophosphohydrolase;
PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
|
| 20 | c2p06A_
|
|
 |
71.2 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af_0060;
PDBTitle: crystal structure of a predicted coding region af_0060 from2 archaeoglobus fulgidus dsm 4304
|
| 21 | d2p06a1 |
|
not modelled |
71.2 |
23 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:AF0060-like |
| 22 | d1ulva4 |
|
not modelled |
41.9 |
11 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Bacterial glucoamylase N-terminal domain-like |
| 23 | d1lf6a2 |
|
not modelled |
38.2 |
21 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Bacterial glucoamylase N-terminal domain-like |
| 24 | d1m4ka2 |
|
not modelled |
33.8 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 25 | d1nkra2 |
|
not modelled |
32.0 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 26 | d1nvma1 |
|
not modelled |
31.1 |
15 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
| 27 | d1qcqa_ |
|
not modelled |
25.1 |
17 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 28 | d1j3ea_ |
|
not modelled |
23.0 |
21 |
Fold:Replication modulator SeqA, C-terminal DNA-binding domain
Superfamily:Replication modulator SeqA, C-terminal DNA-binding domain
Family:Replication modulator SeqA, C-terminal DNA-binding domain |
| 29 | c2a2oG_ |
|
not modelled |
21.7 |
11 |
PDB header:transcription
Chain: G: PDB Molecule:hypothetical protein bt3146;
PDBTitle: crystal structure of a putativetena family transcriptional regulator2 (bt_3146) from bacteroides thetaiotaomicron vpi-5482 at 2.16 a3 resolution
|
| 30 | c2z1dA_ |
|
not modelled |
19.6 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:hydrogenase expression/formation protein hypd;
PDBTitle: crystal structure of [nife] hydrogenase maturation protein, hypd from2 thermococcus kodakaraensis
|
| 31 | d1axib1 |
|
not modelled |
18.1 |
23 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 32 | c2i7uA_ |
|
not modelled |
17.1 |
28 |
PDB header:de novo protein/ligand binding protein
Chain: A: PDB Molecule:four-alpha-helix bundle;
PDBTitle: structural and dynamical analysis of a four-alpha-helix2 bundle with designed anesthetic binding pockets
|
| 33 | d2od6a1 |
|
not modelled |
16.1 |
50 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Marine metagenome family DABB1 |
| 34 | c1z6rC_ |
|
not modelled |
15.6 |
14 |
PDB header:transcription
Chain: C: PDB Molecule:mlc protein;
PDBTitle: crystal structure of mlc from escherichia coli
|
| 35 | c3loeA_ |
|
not modelled |
13.8 |
31 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:neutrophil defensin 1;
PDBTitle: crystal structure of human alpha-defensin 1 (f28a mutant)
|
| 36 | d1z05a2 |
|
not modelled |
13.7 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
| 37 | d1z2ua1 |
|
not modelled |
13.5 |
21 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 38 | d2a2ma1 |
|
not modelled |
13.5 |
11 |
Fold:Heme oxygenase-like
Superfamily:Heme oxygenase-like
Family:TENA/THI-4 |
| 39 | c3gnyA_ |
|
not modelled |
13.3 |
31 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:neutrophil defensin 1;
PDBTitle: crystal structure of human alpha-defensin 1 (hnp1)
|
| 40 | c3lo7A_ |
|
not modelled |
13.2 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:penicillin-binding protein a;
PDBTitle: crystal structure of pbpa from mycobacterium tuberculosis
|
| 41 | d1id3b_ |
|
not modelled |
13.1 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 42 | d1pjua2 |
|
not modelled |
12.8 |
39 |
Fold:Plant proteinase inhibitors
Superfamily:Plant proteinase inhibitors
Family:Plant proteinase inhibitors |
| 43 | c3hj2B_ |
|
not modelled |
12.0 |
31 |
PDB header:antimicrobial protein
Chain: B: PDB Molecule:human neutrophil peptide 1;
PDBTitle: crystal structure of covalent dimer of hnp1
|
| 44 | c3ue3A_ |
|
not modelled |
11.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:septum formation, penicillin binding protein 3,
PDBTitle: crystal structure of acinetobacter baumanni pbp3
|
| 45 | c3equB_ |
|
not modelled |
11.6 |
44 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:penicillin-binding protein 2;
PDBTitle: crystal structure of penicillin-binding protein 2 from neisseria2 gonorrhoeae
|
| 46 | d1nkra1 |
|
not modelled |
11.4 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 47 | d1jeya1 |
|
not modelled |
11.2 |
13 |
Fold:SPOC domain-like
Superfamily:SPOC domain-like
Family:Ku70 subunit middle domain |
| 48 | c2ap1A_ |
|
not modelled |
11.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative regulator protein;
PDBTitle: crystal structure of the putative regulatory protein
|
| 49 | d2zjrs1 |
|
not modelled |
11.0 |
6 |
Fold:Ribosomal protein L25-like
Superfamily:Ribosomal protein L25-like
Family:Ribosomal protein L25-like |
| 50 | c2kloA_ |
|
not modelled |
10.9 |
15 |
PDB header:cell cycle
Chain: A: PDB Molecule:dna replication factor cdt1;
PDBTitle: structure of the cdt1 c-terminal domain
|
| 51 | d2afja1 |
|
not modelled |
10.9 |
9 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:SPRY domain |
| 52 | d1iqpa1 |
|
not modelled |
10.6 |
21 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
| 53 | c1z05A_ |
|
not modelled |
10.5 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, rok family;
PDBTitle: crystal structure of the rok family transcriptional regulator, homolog2 of e.coli mlc protein.
|
| 54 | d1m4ka1 |
|
not modelled |
10.1 |
36 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 55 | c3r4rB_ |
|
not modelled |
9.9 |
25 |
PDB header:cell adhesion
Chain: B: PDB Molecule:hypothetical fimbrial assembly protein;
PDBTitle: crystal structure of a hypothetical fimbrial assembly protein2 (bdi_3522) from parabacteroides distasonis atcc 8503 at 2.38 a3 resolution
|
| 56 | d1z6ra3 |
|
not modelled |
9.9 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
| 57 | c3a4cA_ |
|
not modelled |
9.7 |
16 |
PDB header:cell cycle, replication
Chain: A: PDB Molecule:dna replication factor cdt1;
PDBTitle: crystal structure of cdt1 c terminal domain
|
| 58 | d1ex4a1 |
|
not modelled |
9.6 |
17 |
Fold:SH3-like barrel
Superfamily:DNA-binding domain of retroviral integrase
Family:DNA-binding domain of retroviral integrase |
| 59 | d1rp5a4 |
|
not modelled |
9.2 |
29 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 60 | d1vqqa3 |
|
not modelled |
9.2 |
31 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 61 | d2ayva1 |
|
not modelled |
8.8 |
23 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 62 | d3b5ha2 |
|
not modelled |
8.8 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 63 | d1z3da1 |
|
not modelled |
8.8 |
19 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 64 | c2rkkA_ |
|
not modelled |
8.7 |
19 |
PDB header:lipid transport
Chain: A: PDB Molecule:vacuolar protein sorting-associated protein vta1;
PDBTitle: crystal structure of s.cerevisiae vta1 n-terminal domain
|
| 65 | d2zjrk1 |
|
not modelled |
8.5 |
18 |
Fold:Prokaryotic ribosomal protein L17
Superfamily:Prokaryotic ribosomal protein L17
Family:Prokaryotic ribosomal protein L17 |
| 66 | d2ahme1 |
|
not modelled |
8.4 |
18 |
Fold:Coronavirus NSP8-like
Superfamily:Coronavirus NSP8-like
Family:Coronavirus NSP8-like |
| 67 | d1feua_ |
|
not modelled |
8.4 |
20 |
Fold:Ribosomal protein L25-like
Superfamily:Ribosomal protein L25-like
Family:Ribosomal protein L25-like |
| 68 | c2pmzQ_ |
|
not modelled |
8.4 |
27 |
PDB header:translation, transferase
Chain: Q: PDB Molecule:dna-directed rna polymerase subunit a;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 69 | d2dl2a1 |
|
not modelled |
8.1 |
36 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 70 | d1kx5b_ |
|
not modelled |
8.1 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 71 | d1y6la_ |
|
not modelled |
8.0 |
17 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 72 | d1k38a_ |
|
not modelled |
8.0 |
15 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 73 | c3eo3B_ |
|
not modelled |
7.9 |
16 |
PDB header:isomerase, transferase
Chain: B: PDB Molecule:bifunctional udp-n-acetylglucosamine 2-epimerase/n-
PDBTitle: crystal structure of the n-acetylmannosamine kinase domain of human2 gne protein
|
| 74 | c3ugfB_ |
|
not modelled |
7.7 |
0 |
PDB header:transferase
Chain: B: PDB Molecule:sucrose:(sucrose/fructan) 6-fructosyltransferase;
PDBTitle: crystal structure of a 6-sst/6-sft from pachysandra terminalis
|
| 75 | c3hjlA_ |
|
not modelled |
7.6 |
15 |
PDB header:proton transport
Chain: A: PDB Molecule:flagellar motor switch protein flig;
PDBTitle: the structure of full-length flig from aquifex aeolicus
|
| 76 | c2ahmG_ |
|
not modelled |
7.5 |
18 |
PDB header:viral protein, replication
Chain: G: PDB Molecule:replicase polyprotein 1ab, heavy chain;
PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
|
| 77 | d2huec1 |
|
not modelled |
7.4 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 78 | c2x48B_ |
|
not modelled |
7.2 |
16 |
PDB header:viral protein
Chain: B: PDB Molecule:cag38821;
PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
|
| 79 | d1k47a2 |
|
not modelled |
7.2 |
10 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Phosphomevalonate kinase (PMK) |
| 80 | c3tixA_ |
|
not modelled |
7.1 |
19 |
PDB header:gene regulation/protein binding
Chain: A: PDB Molecule:ubiquitin-like protein smt3, rna-induced transcriptional
PDBTitle: crystal structure of the chp1-tas3 complex core
|
| 81 | c1mwuA_ |
|
not modelled |
7.1 |
31 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:penicillin-binding protein 2a;
PDBTitle: structure of methicillin acyl-penicillin binding protein 2a2 from methicillin resistant staphylococcus aureus strain3 27r at 2.60 a resolution.
|
| 82 | d1w1we_ |
|
not modelled |
7.1 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Rad21/Rec8-like |
| 83 | c1w1wF_ |
|
not modelled |
7.1 |
29 |
PDB header:cell adhesion
Chain: F: PDB Molecule:sister chromatid cohesion protein 1;
PDBTitle: sc smc1hd:scc1-c complex, atpgs
|
| 84 | d2aaka_ |
|
not modelled |
7.0 |
15 |
Fold:UBC-like
Superfamily:UBC-like
Family:UBC-related |
| 85 | d1cg2a2 |
|
not modelled |
7.0 |
13 |
Fold:Ferredoxin-like
Superfamily:Bacterial exopeptidase dimerisation domain
Family:Bacterial exopeptidase dimerisation domain |
| 86 | c2jtdA_ |
|
not modelled |
7.0 |
10 |
PDB header:cell adhesion
Chain: A: PDB Molecule:myomesin-1;
PDBTitle: skelemin immunoglobulin c2 like domain 4
|
| 87 | d1ov9a_ |
|
not modelled |
7.0 |
35 |
Fold:H-NS histone-like proteins
Superfamily:H-NS histone-like proteins
Family:H-NS histone-like proteins |
| 88 | d1ryqa_ |
|
not modelled |
6.9 |
42 |
Fold:Rubredoxin-like
Superfamily:RNA polymerase subunits
Family:RpoE2-like |
| 89 | d2ok3a1 |
|
not modelled |
6.9 |
24 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:Cyclophilin (peptidylprolyl isomerase) |
| 90 | d1kg1a_ |
|
not modelled |
6.9 |
33 |
Fold:Necrosis inducing protein 1, NIP1
Superfamily:Necrosis inducing protein 1, NIP1
Family:Necrosis inducing protein 1, NIP1 |
| 91 | c2z56B_ |
|
not modelled |
6.9 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:tk-subtilisin;
PDBTitle: crystal structure of g56s-propeptide:s324a-subtilisin complex
|
| 92 | c3oc2A_ |
|
not modelled |
6.8 |
19 |
PDB header:penicillin-binding protein
Chain: A: PDB Molecule:penicillin-binding protein 3;
PDBTitle: crystal structure of penicillin-binding protein 3 from pseudomonas2 aeruginosa
|
| 93 | d2fnja1 |
|
not modelled |
6.8 |
18 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:SPRY domain |
| 94 | c2wadB_ |
|
not modelled |
6.7 |
18 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:penicillin-binding protein 2b;
PDBTitle: penicillin-binding protein 2b (pbp-2b) from streptococcus2 pneumoniae (strain 5204)
|
| 95 | d2f09a1 |
|
not modelled |
6.6 |
19 |
Fold:Streptavidin-like
Superfamily:YdhA-like
Family:YdhA-like |
| 96 | d1k25a4 |
|
not modelled |
6.5 |
29 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 97 | d1dfup_ |
|
not modelled |
6.5 |
9 |
Fold:Ribosomal protein L25-like
Superfamily:Ribosomal protein L25-like
Family:Ribosomal protein L25-like |
| 98 | d2z3qa1 |
|
not modelled |
6.4 |
28 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Short-chain cytokines |
| 99 | d1pyya4 |
|
not modelled |
6.4 |
29 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |