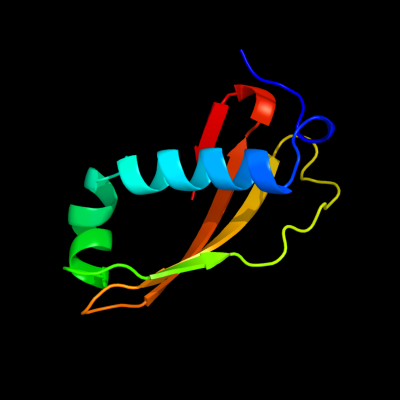
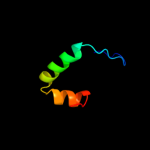
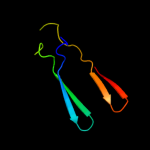
1 c2xglB_
100.0
26
PDB header: antibioticChain: B: PDB Molecule: colicin-m immunity protein;PDBTitle: the x-ray structure of the escherichia coli colicin m immunity2 protein demonstrates the presence of a disulphide bridge, which is3 functionally essential
2 c3dd7A_
47.1
13
PDB header: ribosome inhibitorChain: A: PDB Molecule: death on curing protein;PDBTitle: structure of doch66y in complex with the c-terminal domain of phd
3 d2nxyb2
38.8
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C2 set domains4 c3fsnA_
25.1
20
PDB header: isomeraseChain: A: PDB Molecule: retinal pigment epithelium-specific 65 kda protein;PDBTitle: crystal structure of rpe65 at 2.14 angstrom resolution
5 c2k87A_
21.2
24
PDB header: viral protein, rna binding proteinChain: A: PDB Molecule: non-structural protein 3 of replicase polyprotein 1a;PDBTitle: nmr structure of a putative rna binding protein (sars1) from sars2 coronavirus
6 c2qh7A_
19.3
40
PDB header: metal binding proteinChain: A: PDB Molecule: zinc finger cdgsh-type domain 1;PDBTitle: mitoneet is a uniquely folded 2fe-2s outer mitochondrial membrane2 protein stabilized by pioglitazone
7 c3fnvB_
18.8
50
PDB header: metal binding proteinChain: B: PDB Molecule: cdgsh iron sulfur domain-containing protein 2;PDBTitle: crystal structure of miner1: the redox-active 2fe-2s protein causative2 in wolfram syndrome 2
8 d1wh6a_
16.2
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain9 c2l25A_
15.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: np_888769.1
10 c3iynR_
14.4
40
PDB header: virusChain: R: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
11 c3arcl_
14.3
33
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
12 d1s7ea2
13.6
6
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain13 c3kfoA_
13.1
17
PDB header: protein transportChain: A: PDB Molecule: nucleoporin nup133;PDBTitle: crystal structure of the c-terminal domain from the nuclear pore2 complex component nup133 from saccharomyces cerevisiae
14 d2csfa1
13.0
8
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain15 d1o6ea_
12.8
21
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin16 d1r17a1
12.1
12
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Fibrinogen-binding domain17 d1fyhb2
11.3
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III18 c2l9wA_
10.4
12
PDB header: splicing, rna binding proteinChain: A: PDB Molecule: u4/u6 snrna-associated-splicing factor prp24;PDBTitle: solution structure of the c-terminal domain of prp24
19 d1iega_
10.4
23
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin20 c3nqyA_
9.8
18
PDB header: hydrolaseChain: A: PDB Molecule: secreted metalloprotease mcp02;PDBTitle: crystal structure of the autoprocessed complex of vibriolysin mcp-022 with a single point mutation e346a
21 d2pbka1
not modelled
9.7
13
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin22 c2j0kB_
not modelled
9.3
20
PDB header: transferaseChain: B: PDB Molecule: focal adhesion kinase 1;PDBTitle: crystal structure of a fragment of focal adhesion kinase2 containing the ferm and kinase domains.
23 d1at3a_
not modelled
9.2
16
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin24 c2eo1A_
not modelled
9.0
22
PDB header: contractile proteinChain: A: PDB Molecule: cdna flj14124 fis, clone mamma1002498;PDBTitle: solution structure of the ig domain of human obscn protein
25 d1wh8a_
not modelled
8.5
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain26 d1x2la1
not modelled
8.1
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain27 c2druA_
not modelled
7.9
8
PDB header: immune systemChain: A: PDB Molecule: chimera of cd48 antigen and t-cell surface antigen cd2;PDBTitle: crystal structure and binding properties of the cd2 and cd244 (2b4)2 binding protein, cd48
28 c3p02A_
not modelled
7.8
21
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein of unknown function (bacova_00267) from2 bacteroides ovatus at 1.55 a resolution
29 d1vcaa2
not modelled
7.6
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains30 d1biha3
not modelled
7.5
5
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains31 c2yqeA_
not modelled
6.8
22
PDB header: dna binding proteinChain: A: PDB Molecule: jumonji/arid domain-containing protein 1d;PDBTitle: solution structure of the arid domain of jarid1d protein
32 c2axtl_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
33 c3a0bL_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
34 c3arcL_
not modelled
6.5
33
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
35 c3a0bl_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
36 c3bz1L_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
37 c2axtL_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
38 c3prrL_
not modelled
6.5
33
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
39 c3bz2L_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
40 c1s5lL_
not modelled
6.5
33
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
41 c3prqL_
not modelled
6.5
33
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
42 c3kziL_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
43 c3a0hl_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
44 d2axtl1
not modelled
6.5
33
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like45 c1s5ll_
not modelled
6.5
33
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
46 c3a0hL_
not modelled
6.5
33
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
47 c3h6sE_
not modelled
6.5
18
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: clitocypin analog;PDBTitle: strucure of clitocypin - cathepsin v complex
48 c2dkuA_
not modelled
6.3
15
PDB header: contractile proteinChain: A: PDB Molecule: kiaa1556 protein;PDBTitle: solution structure of the third ig-like domain of human2 kiaa1556 protein
49 c3nqkA_
not modelled
6.3
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a structural genomics, unknown function2 (bacova_03322) from bacteroides ovatus at 2.61 a resolution
50 c3ts7B_
not modelled
6.0
17
PDB header: transferaseChain: B: PDB Molecule: geranyltranstransferase;PDBTitle: crystal structure of farnesyl diphosphate synthase (target efi-501951)2 from methylococcus capsulatus
51 c3epzA_
not modelled
5.8
15
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: structure of the replication foci-targeting sequence of human dna2 cytosine methyltransferase dnmt1
52 d1tdpa_
not modelled
5.7
14
Fold: Bromodomain-likeSuperfamily: Bacteriocin immunity protein-likeFamily: Carnobacteriocin B2 immunity protein53 d2cs7a1
not modelled
5.6
38
Fold: IL8-likeSuperfamily: PhtA domain-likeFamily: PhtA domain-like54 c2q97T_
not modelled
5.4
24
PDB header: structural protein/cell invasionChain: T: PDB Molecule: toxofilin;PDBTitle: complex of mammalian actin with toxofilin from toxoplasma gondii
55 c3rjoA_
not modelled
5.4
24
PDB header: hydrolaseChain: A: PDB Molecule: endoplasmic reticulum aminopeptidase 1;PDBTitle: crystal structure of erap1 peptide binding domain
56 d1gpja3
not modelled
5.2
25
Fold: Ferredoxin-likeSuperfamily: Glutamyl tRNA-reductase catalytic, N-terminal domainFamily: Glutamyl tRNA-reductase catalytic, N-terminal domain57 d1ni5a3
not modelled
5.2
17
Fold: PheT/TilS domainSuperfamily: PheT/TilS domainFamily: tRNA-Ile-lysidine synthetase, TilS, C-terminal domain58 c2cpcA_
not modelled
5.2
11
PDB header: immune systemChain: A: PDB Molecule: kiaa0657 protein;PDBTitle: solution structure of rsgi ruh-030, an ig like domain from2 human cdna