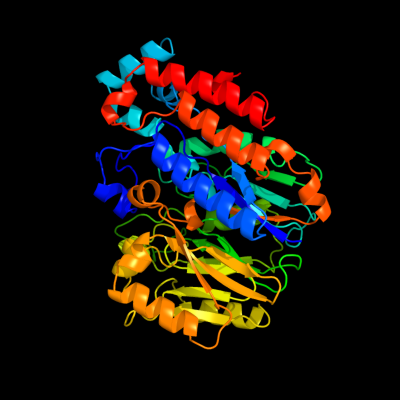
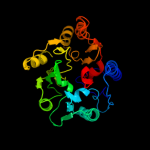
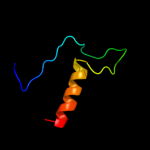
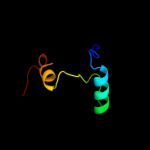
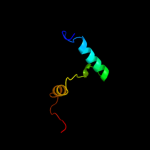
1 d1ynha1
100.0
100
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Succinylarginine dihydrolase-like2 d1h70a_
97.0
19
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Dimethylarginine dimethylaminohydrolase DDAH3 d1bwda_
90.3
12
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Amidinotransferase4 c3dydB_
81.8
18
PDB header: transferaseChain: B: PDB Molecule: tyrosine aminotransferase;PDBTitle: human tyrosine aminotransferase
5 d1jdwa_
76.1
15
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Amidinotransferase6 c1jdwA_
76.1
15
PDB header: transferaseChain: A: PDB Molecule: l-arginine\:glycine amidinotransferase;PDBTitle: crystal structure and mechanism of l-arginine: glycine2 amidinotransferase: a mitochondrial enzyme involved in3 creatine biosynthesis
7 c2ci6A_
75.9
19
PDB header: hydrolaseChain: A: PDB Molecule: ng, ng-dimethylarginine dimethylaminohydrolase 1;PDBTitle: crystal structure of dimethylarginine2 dimethylaminohydrolase i bound with zinc low ph
8 d1wsta1
66.4
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like9 c8jdwA_
64.6
15
PDB header: transferaseChain: A: PDB Molecule: protein (l-arginine:glycine amidinotransferase);PDBTitle: crystal structure of human l-arginine:glycine2 amidinotransferase in complex with l-alanine
10 c3i4aA_
61.2
18
PDB header: hydrolaseChain: A: PDB Molecule: n(g),n(g)-dimethylarginine dimethylaminohydrolasePDBTitle: crystal structure of dimethylarginine2 dimethylaminohydrolase-1 (ddah-1) in complex with n5-(1-3 iminopropyl)-l-ornithine
11 d1bl0a1
53.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator12 c3pdxA_
53.4
19
PDB header: transferaseChain: A: PDB Molecule: tyrosine aminotransferase;PDBTitle: crystal structural of mouse tyrosine aminotransferase
13 d1d5ya1
43.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator14 c3fkdC_
41.8
15
PDB header: lyaseChain: C: PDB Molecule: l-threonine-o-3-phosphate decarboxylase;PDBTitle: the crystal structure of l-threonine-o-3-phosphate2 decarboxylase from porphyromonas gingivalis
15 c3p1tB_
37.3
13
PDB header: transferaseChain: B: PDB Molecule: putative histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a putative aminotransferase (bpsl1724) from2 burkholderia pseudomallei k96243 at 2.60 a resolution
16 c3ctzA_
36.7
19
PDB header: hydrolaseChain: A: PDB Molecule: xaa-pro aminopeptidase 1;PDBTitle: structure of human cytosolic x-prolyl aminopeptidase
17 d2bo9b1
32.8
25
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Latexin-like18 c3eibB_
31.2
12
PDB header: transferaseChain: B: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: crystal structure of k270n variant of ll-diaminopimelate2 aminotransferase from arabidopsis thaliana
19 c3oouA_
30.0
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2118 protein;PDBTitle: the structure of a protein with unkown function from listeria innocua
20 c3mklB_
28.6
11
PDB header: transcription regulatorChain: B: PDB Molecule: hth-type transcriptional regulator gadx;PDBTitle: crystal structure of dna-binding transcriptional dual regulator from2 escherichia coli k-12
21 d1rxxa_
not modelled
28.3
24
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Arginine deiminase22 d1fg7a_
not modelled
28.0
30
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like23 c1bl0A_
not modelled
27.8
12
PDB header: transcription/dnaChain: A: PDB Molecule: protein (multiple antibiotic resistance protein);PDBTitle: multiple antibiotic resistance protein (mara)/dna complex
24 c3t18D_
not modelled
26.0
16
PDB header: transferaseChain: D: PDB Molecule: aminotransferase class i and ii;PDBTitle: crystal structure of aminotransferase from anaerococcus prevotii dsm2 20548.
25 c3g7qA_
not modelled
24.7
12
PDB header: transferaseChain: A: PDB Molecule: valine-pyruvate aminotransferase;PDBTitle: crystal structure of valine-pyruvate aminotransferase avta2 (np_462565.1) from salmonella typhimurium lt2 at 1.80 a resolution
26 c2k9sA_
not modelled
22.1
15
PDB header: transcriptionChain: A: PDB Molecule: arabinose operon regulatory protein;PDBTitle: solution structure of dna binding domain of e. coli arac
27 c2of5K_
not modelled
21.7
15
PDB header: apoptosisChain: K: PDB Molecule: leucine-rich repeat and death domain-containingPDBTitle: oligomeric death domain complex
28 c3hdoB_
not modelled
21.6
19
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
29 c3oioA_
not modelled
21.0
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (arac-type dna-binding domain-PDBTitle: crystal structure of transcriptional regulator (arac-type dna-binding2 domain-containing proteins) from chromobacterium violaceum
30 d2v7fa1
not modelled
19.0
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rps19E-like31 d2vnud3
not modelled
18.5
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like32 c2bo9B_
not modelled
18.4
25
PDB header: hydrolaseChain: B: PDB Molecule: human latexin;PDBTitle: human carboxypeptidase a4 in complex with human latexin.
33 d2foka1
not modelled
18.4
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Restriction endonuclease FokI, N-terminal (recognition) domain34 c3e4wB_
not modelled
17.6
28
PDB header: oxidoreductaseChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a 33kda catalase-related protein from2 mycobacterium avium subsp. paratuberculosis. p2(1)2(1)2(1) crystal3 form.
35 c3nzlA_
not modelled
17.4
35
PDB header: transcriptionChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: crystal structure of the n-terminal domain of dna-binding protein2 satb1 from homo sapiens, northeast structural genomics consortium3 target hr4435b
36 c3gbgA_
not modelled
17.3
12
PDB header: transcription regulatorChain: A: PDB Molecule: tcp pilus virulence regulatory protein;PDBTitle: crystal structure of toxt from vibrio cholerae o395
37 c3iz6S_
not modelled
16.5
20
PDB header: ribosomeChain: S: PDB Molecule: 40s ribosomal protein s19 (s19e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
38 c2zc0C_
not modelled
15.8
17
PDB header: transferaseChain: C: PDB Molecule: alanine glyoxylate transaminase;PDBTitle: crystal structure of an archaeal alanine:glyoxylate aminotransferase
39 d1s9ra_
not modelled
15.5
12
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Arginine deiminase40 d1lc5a_
not modelled
15.5
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like41 d1x0ma1
not modelled
14.7
9
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like42 c3jyvT_
not modelled
14.6
20
PDB header: ribosomeChain: T: PDB Molecule: s19e protein;PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
43 d1vp4a_
not modelled
14.6
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like44 c3mn2B_
not modelled
14.5
8
PDB header: transcription regulatorChain: B: PDB Molecule: probable arac family transcriptional regulator;PDBTitle: the crystal structure of a probable arac family transcriptional2 regulator from rhodopseudomonas palustris cga009
45 c3cq6E_
not modelled
13.9
17
PDB header: transferaseChain: E: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: histidinol-phosphate aminotransferase from corynebacterium2 glutamicum holo-form (plp covalently bound )
46 c3ismA_
not modelled
13.6
19
PDB header: hydrolase inhibitor/hydrolaseChain: A: PDB Molecule: cg8862;PDBTitle: crystal structure of the endog/endogi complex: mechanism of endog2 inhibition
47 c3getA_
not modelled
13.4
22
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (np_281508.1) from campylobacter jejuni at 2.01 a resolution
48 c3ftbA_
not modelled
13.2
10
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: the crystal structure of the histidinol-phosphate2 aminotransferase from clostridium acetobutylicum
49 c3aiiA_
not modelled
12.7
26
PDB header: ligaseChain: A: PDB Molecule: glutamyl-trna synthetase;PDBTitle: archaeal non-discriminating glutamyl-trna synthetase from2 methanothermobacter thermautotrophicus
50 c3by0B_
not modelled
12.5
18
PDB header: ligand binding proteinChain: B: PDB Molecule: neutrophil gelatinase-associated lipocalin;PDBTitle: crystal structure of siderocalin (ngal, lipocalin 2) w79a-r81a2 complexed with ferric enterobactin
51 c3lsgD_
not modelled
12.4
18
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
52 c3ly1C_
not modelled
12.0
16
PDB header: transferaseChain: C: PDB Molecule: putative histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (yp_050345.1) from erwinia carotovora atroseptica scri1043 at 1.80 a3 resolution
53 c3ezsB_
not modelled
11.8
13
PDB header: transferaseChain: B: PDB Molecule: aminotransferase aspb;PDBTitle: crystal structure of aminotransferase aspb (np_207418.1) from2 helicobacter pylori 26695 at 2.19 a resolution
54 d1bw0a_
not modelled
11.7
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like55 c3s1sA_
not modelled
11.4
10
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
56 c2douA_
not modelled
10.9
18
PDB header: transferaseChain: A: PDB Molecule: probable n-succinyldiaminopimelate aminotransferase;PDBTitle: probable n-succinyldiaminopimelate aminotransferase (ttha0342) from2 thermus thermophilus hb8
57 c2re3A_
not modelled
10.6
45
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1285 family protein (spo_0140) from2 silicibacter pomeroyi dss-3 at 2.50 a resolution
58 c2xznT_
not modelled
10.5
17
PDB header: ribosomeChain: T: PDB Molecule: rps19e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
59 c2yx5A_
not modelled
10.1
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0062 protein mj1593;PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
60 d1n26a1
not modelled
10.0
35
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains61 d2nwva1
not modelled
9.9
39
Fold: XisI-likeSuperfamily: XisI-likeFamily: XisI-like62 d1v30a_
not modelled
9.8
43
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like63 c2e12B_
not modelled
9.7
50
PDB header: translationChain: B: PDB Molecule: hypothetical protein xcc3642;PDBTitle: the crystal structure of xc5848 from xanthomonas campestris2 adopting a novel variant of sm-like motif
64 d1xhsa_
not modelled
9.6
71
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like65 c1d5yD_
not modelled
9.5
15
PDB header: transcription/dnaChain: D: PDB Molecule: rob transcription factor;PDBTitle: crystal structure of the e. coli rob transcription factor2 in complex with dna
66 c2wo3B_
not modelled
9.4
40
PDB header: transferase/signaling proteinChain: B: PDB Molecule: ephrin-a2;PDBTitle: crystal structure of the epha4-ephrina2 complex
67 c3ke2A_
not modelled
9.4
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yp_928783.1;PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
68 c2k6lA_
not modelled
9.0
50
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
69 c3a0hL_
not modelled
8.8
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
70 c3a0hl_
not modelled
8.8
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
71 d2nlva1
not modelled
8.8
28
Fold: XisI-likeSuperfamily: XisI-likeFamily: XisI-like72 c3if2B_
not modelled
8.7
14
PDB header: transferaseChain: B: PDB Molecule: aminotransferase;PDBTitle: crystal structure of putative amino-acid aminotransferase2 (yp_265399.1) from psychrobacter arcticum 273-4 at 2.50 a resolution
73 c3arcL_
not modelled
8.6
67
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
74 c1s5lL_
not modelled
8.6
67
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
75 c3prrL_
not modelled
8.6
67
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
76 c2axtl_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
77 c3a0bL_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
78 c3a0bl_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
79 c3bz1L_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
80 d2axtl1
not modelled
8.6
67
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like81 c3bz2L_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
82 c3prqL_
not modelled
8.6
67
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
83 c2axtL_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
84 c3kziL_
not modelled
8.6
67
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
85 c1s5ll_
not modelled
8.6
67
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
86 c2e58D_
not modelled
8.5
22
PDB header: transferaseChain: D: PDB Molecule: mnmc2;PDBTitle: crystal structure of mnmc2 from aquifex aeolicus
87 d1j32a_
not modelled
8.2
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like88 c2euzA_
not modelled
7.9
29
PDB header: cell cycle/dnaChain: A: PDB Molecule: ndt80 protein;PDBTitle: structure of a ndt80-dna complex (mse mutant mc5t)
89 d1mnna_
not modelled
7.6
29
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: DNA-binding domain from NDT8090 d1dqna_
not modelled
7.5
32
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)91 c1wr1B_
not modelled
7.5
29
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like protein dsk2;PDBTitle: the complex sturcture of dsk2p uba with ubiquitin
92 d1xi9a_
not modelled
7.2
11
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like93 d2p97a1
not modelled
7.0
15
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Ava3068-like94 d1smye_
not modelled
6.9
41
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RNA polymerase omega subunit95 c3ol4B_
not modelled
6.7
15
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
96 c2xstA_
not modelled
6.6
13
PDB header: transport proteinChain: A: PDB Molecule: lipocalin 15;PDBTitle: crystal structure of the human lipocalin 15
97 d1dxha2
not modelled
6.6
8
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase98 d1lfpa_
not modelled
6.5
16
Fold: YebC-likeSuperfamily: YebC-likeFamily: YebC-like99 d1gg4a3
not modelled
6.4
9
Fold: MurF and HprK N-domain-likeSuperfamily: MurE/MurF N-terminal domainFamily: MurE/MurF N-terminal domain