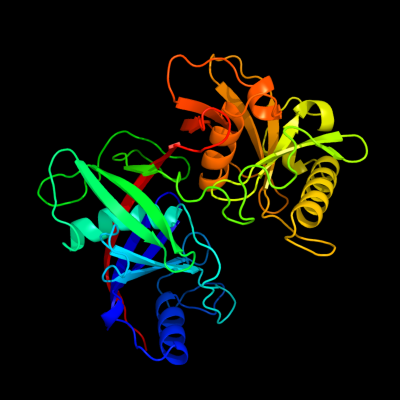
| 1 | c3g7kD_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: D: PDB Molecule:3-methylitaconate isomerase;
PDBTitle: crystal structure of methylitaconate-delta-isomerase
|
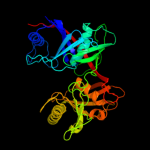
| 2 | c2pw0A_
|
|
 |
100.0 |
39 |
PDB header:unknown function
Chain: A: PDB Molecule:prpf methylaconitate isomerase;
PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
|
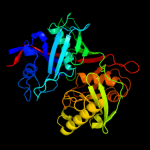
| 3 | d2h9fa1
|
|
 |
100.0 |
42 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PA0793-like |
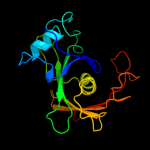
| 4 | d2h9fa2
|
|
 |
100.0 |
30 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PA0793-like |
| 5 | c1w62B_
|
|
 |
99.4 |
18 |
PDB header:racemase
Chain: B: PDB Molecule:b-cell mitogen;
PDBTitle: proline racemase in complex with one molecule of pyrrole-2-2 carboxylic acid (hemi form)
|
| 6 | d1tm0a_
|
|
 |
98.5 |
26 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Proline racemase |
| 7 | c2gkjA_
|
|
 |
98.4 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase in complex2 with an irreversible inhibitor dl-azidap
|
| 8 | c3ednB_
|
|
 |
98.1 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:phenazine biosynthesis protein, phzf family;
PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
|
| 9 | c3ekmE_
|
|
 |
98.0 |
16 |
PDB header:isomerase
Chain: E: PDB Molecule:diaminopimelate epimerase, chloroplastic;
PDBTitle: crystal structure of diaminopimelate epimerase form2 arabidopsis thaliana in complex with irreversible inhibitor3 dl-azidap
|
| 10 | c1ym5A_
|
|
 |
97.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical 32.6 kda protein in dap2-slt2
PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
|
| 11 | d1xuba1
|
|
 |
97.3 |
18 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 12 | c1u1wA_
|
|
 |
97.2 |
18 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:phenazine biosynthesis protein phzf;
PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
|
| 13 | d1qy9a1
|
|
 |
96.4 |
18 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 14 | d1s7ja_
|
|
 |
95.9 |
13 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 15 | c1qy9B_
|
|
 |
95.6 |
16 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ydde;
PDBTitle: crystal structure of e. coli se-met protein ydde
|
| 16 | d1xuba2
|
|
 |
95.2 |
18 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 17 | d1qy9a2
|
|
 |
95.2 |
16 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 18 | c3fveA_
|
|
 |
95.1 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
|
| 19 | c2otnB_
|
|
 |
94.8 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
|
| 20 | d1u0ka1
|
|
 |
93.4 |
10 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 21 | d2gkea1 |
|
not modelled |
93.3 |
17 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
| 22 | d2gkea2 |
|
not modelled |
90.2 |
16 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
| 23 | c2azpA_ |
|
not modelled |
87.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa1268;
PDBTitle: crystal structure of pa1268 solved by sulfur sad
|
| 24 | c1u0kA_ |
|
not modelled |
83.8 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:gene product pa4716;
PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
|
| 25 | c1pyuD_ |
|
not modelled |
61.1 |
21 |
PDB header:lyase
Chain: D: PDB Molecule:aspartate 1-decarboxylase alfa chain;
PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
|
| 26 | c3ougA_ |
|
not modelled |
58.5 |
36 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
|
| 27 | c1vc3B_ |
|
not modelled |
57.5 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:l-aspartate-alpha-decarboxylase heavy chain;
PDBTitle: crystal structure of l-aspartate-alpha-decarboxylase
|
| 28 | c1pt1B_ |
|
not modelled |
56.1 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
|
| 29 | d1ppya_ |
|
not modelled |
55.7 |
21 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Pyruvoyl dependent aspartate decarboxylase, ADC |
| 30 | c2c45F_ |
|
not modelled |
51.5 |
32 |
PDB header:lyase
Chain: F: PDB Molecule:aspartate 1-decarboxylase precursor;
PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
|
| 31 | d1bk8a_ |
|
not modelled |
39.2 |
31 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Plant defensins |
| 32 | d1yq2a1 |
|
not modelled |
36.4 |
13 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 33 | d1kl7a_ |
|
not modelled |
25.5 |
44 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 34 | d1jz8a2 |
|
not modelled |
25.1 |
12 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 35 | d1h6ua1 |
|
not modelled |
24.7 |
32 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Internalin Ig-like domain |
| 36 | d2omza1 |
|
not modelled |
23.6 |
26 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Internalin Ig-like domain |
| 37 | c3opwA_ |
|
not modelled |
21.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dna damage-responsive transcriptional repressor rph1;
PDBTitle: crystal structure of the rph1 catalytic core
|
| 38 | d1h6ta1 |
|
not modelled |
21.2 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Internalin Ig-like domain |
| 39 | c2wh7A_ |
|
not modelled |
17.5 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronidase-phage associated;
PDBTitle: the partial structure of a group a streptpcoccal phage-2 encoded tail fibre hyaluronate lyase hylp2
|
| 40 | d1w5sa1 |
|
not modelled |
16.7 |
46 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
| 41 | d1xhja_ |
|
not modelled |
14.3 |
18 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 42 | d1ayja_ |
|
not modelled |
13.8 |
23 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Plant defensins |
| 43 | c4proD_ |
|
not modelled |
12.8 |
22 |
PDB header:serine protease
Chain: D: PDB Molecule:alpha-lytic protease;
PDBTitle: alpha-lytic protease complexed with pro region
|
| 44 | d1yuaa1 |
|
not modelled |
12.7 |
29 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Prokaryotic DNA topoisomerase I, a C-terminal fragment |
| 45 | d1xpma2 |
|
not modelled |
12.4 |
27 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 46 | c2qj8B_ |
|
not modelled |
12.4 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:mlr6093 protein;
PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
|
| 47 | d1ee0a2 |
|
not modelled |
12.2 |
36 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 48 | d1u0ka2 |
|
not modelled |
12.0 |
23 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
| 49 | c1yuaA_ |
|
not modelled |
11.1 |
29 |
PDB header:dna binding protein
Chain: A: PDB Molecule:topoisomerase i;
PDBTitle: c-terminal domain of escherichia coli topoisomerase i
|
| 50 | c3tsyA_ |
|
not modelled |
11.0 |
31 |
PDB header:ligase, transferase
Chain: A: PDB Molecule:fusion protein 4-coumarate--coa ligase 1, resveratrol
PDBTitle: 4-coumaroyl-coa ligase::stilbene synthase fusion protein
|
| 51 | c3muwU_ |
|
not modelled |
10.9 |
15 |
PDB header:virus
Chain: U: PDB Molecule:structural polyprotein;
PDBTitle: pseudo-atomic structure of the e2-e1 protein shell in sindbis virus
|
| 52 | d1bi5a2 |
|
not modelled |
10.3 |
36 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 53 | d1u0ua2 |
|
not modelled |
10.3 |
36 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 54 | d1xafa_ |
|
not modelled |
9.7 |
20 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:YfiH-like |
| 55 | d1gpqa_ |
|
not modelled |
9.1 |
36 |
Fold:Inhibitor of vertebrate lysozyme, Ivy
Superfamily:Inhibitor of vertebrate lysozyme, Ivy
Family:Inhibitor of vertebrate lysozyme, Ivy |
| 56 | d1tbxa_ |
|
not modelled |
8.9 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:F93-like |
| 57 | d2b59b2 |
|
not modelled |
8.5 |
33 |
Fold:Prealbumin-like
Superfamily:Carboxypeptidase regulatory domain-like
Family:Pre-dockerin domain |
| 58 | c2klxA_ |
|
not modelled |
8.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin;
PDBTitle: solution structure of glutaredoxin from bartonella henselae str.2 houston
|
| 59 | c2bs5A_ |
|
not modelled |
8.2 |
29 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:fucose-binding lectin protein;
PDBTitle: lectin from ralstonia solanacearum complexed with 2-2 fucosyllactose
|
| 60 | c1uheA_ |
|
not modelled |
8.0 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase alpha chain;
PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
|
| 61 | c3a5iB_ |
|
not modelled |
7.6 |
37 |
PDB header:protein transport
Chain: B: PDB Molecule:flagellar biosynthesis protein flha;
PDBTitle: structure of the cytoplasmic domain of flha
|
| 62 | d1rv9a_ |
|
not modelled |
7.5 |
20 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:YfiH-like |
| 63 | c3mixA_ |
|
not modelled |
7.5 |
45 |
PDB header:protein transport
Chain: A: PDB Molecule:flagellar biosynthesis protein flha;
PDBTitle: crystal structure of the cytosolic domain of b. subtilis flha
|
| 64 | c2d3mA_ |
|
not modelled |
7.1 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:pentaketide chromone synthase;
PDBTitle: pentaketide chromone synthase complexed with coenzyme a
|
| 65 | c1txtB_ |
|
not modelled |
7.0 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:3-hydroxy-3-methylglutaryl-coa synthase;
PDBTitle: staphylococcus aureus 3-hydroxy-3-methylglutaryl-coa2 synthase
|
| 66 | d1gmia_ |
|
not modelled |
6.8 |
11 |
Fold:C2 domain-like
Superfamily:C2 domain (Calcium/lipid-binding domain, CaLB)
Family:PLC-like (P variant) |
| 67 | d2ckca1 |
|
not modelled |
6.6 |
35 |
Fold:GYF/BRK domain-like
Superfamily:BRK domain-like
Family:BRK domain-like |
| 68 | c2ckcA_ |
|
not modelled |
6.6 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:chromodomain-helicase-dna-binding protein 7;
PDBTitle: solution structures of the brk domains of the human chromo2 helicase domain 7 and 8, reveals structural similarity3 with gyf domain suggesting a role in protein interaction
|
| 69 | c3o6xC_ |
|
not modelled |
6.6 |
20 |
PDB header:ligase
Chain: C: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of the type iii glutamine synthetase from2 bacteroides fragilis
|
| 70 | d1ueba1 |
|
not modelled |
6.3 |
19 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 71 | d1t8ha_ |
|
not modelled |
6.2 |
25 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:YfiH-like |
| 72 | d1u0ma2 |
|
not modelled |
6.2 |
31 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 73 | c2ht9A_ |
|
not modelled |
6.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin-2;
PDBTitle: the structure of dimeric human glutaredoxin 2
|
| 74 | d2yvxa3 |
|
not modelled |
5.9 |
19 |
Fold:MgtE membrane domain-like
Superfamily:MgtE membrane domain-like
Family:MgtE membrane domain-like |
| 75 | c3brcA_ |
|
not modelled |
5.9 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein of unknown function;
PDBTitle: crystal structure of a conserved protein of unknown function from2 methanobacterium thermoautotrophicum
|
| 76 | c2xfbI_ |
|
not modelled |
5.9 |
20 |
PDB header:virus
Chain: I: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: the chikungunya e1 e2 envelope glycoprotein complex fit into2 the sindbis virus cryo-em map
|
| 77 | c2zunB_ |
|
not modelled |
5.9 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:458aa long hypothetical endo-1,4-beta-glucanase;
PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii
|
| 78 | d2v0ea1 |
|
not modelled |
5.8 |
35 |
Fold:GYF/BRK domain-like
Superfamily:BRK domain-like
Family:BRK domain-like |
| 79 | c3izcG_ |
|
not modelled |
5.8 |
17 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein rpl6 (l6e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 80 | c3n43B_ |
|
not modelled |
5.8 |
20 |
PDB header:viral protein
Chain: B: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: crystal structures of the mature envelope glycoprotein complex2 (trypsin cleavage) of chikungunya virus.
|
| 81 | c3n40P_ |
|
not modelled |
5.7 |
18 |
PDB header:viral protein
Chain: P: PDB Molecule:p62 envelope glycoprotein;
PDBTitle: crystal structure of the immature envelope glycoprotein complex of2 chikungunya virus.
|
| 82 | d1rw0a_ |
|
not modelled |
5.5 |
23 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:YfiH-like |
| 83 | d1b24a2 |
|
not modelled |
5.3 |
43 |
Fold:Homing endonuclease-like
Superfamily:Homing endonucleases
Family:Group I mobile intron endonuclease |
| 84 | d1a77a1 |
|
not modelled |
5.2 |
23 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 85 | c3mydA_ |
|
not modelled |
5.1 |
32 |
PDB header:protein transport
Chain: A: PDB Molecule:flagellar biosynthesis protein flha;
PDBTitle: structure of the cytoplasmic domain of flha from helicobacter pylori
|
| 86 | c2xm5A_ |
|
not modelled |
5.1 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:cloq;
PDBTitle: structural and mechanistic analysis of the magnesium-2 independent aromatic prenyltransferase cloq from the3 clorobiocin biosynthetic pathway
|
| 87 | c2p8uB_ |
|
not modelled |
5.1 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa synthase, cytoplasmic;
PDBTitle: crystal structure of human 3-hydroxy-3-methylglutaryl coa synthase i
|
| 88 | c3lehA_ |
|
not modelled |
5.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative hydroxymethylglutaryl-coa synthase;
PDBTitle: the crystal structure of smu.943c from streptococcus mutans ua159
|