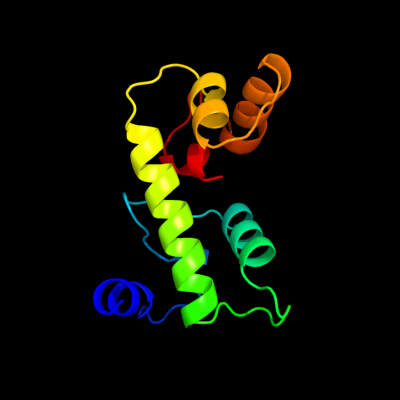
1 c3sibA_
86.6
13
PDB header: dna binding proteinChain: A: PDB Molecule: ure3-bp sequence specific dna binding protein;PDBTitle: crystal structure of ure3-binding protein, wild-type
2 d1brwa1
56.9
12
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain3 d2tpta1
48.9
17
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain4 c3gh1A_
48.4
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted nucleotide-binding protein;PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
5 c3bq9A_
48.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted rossmann fold nucleotide-binding domain-PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
6 d1ldda_
44.4
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: SCF ubiquitin ligase complex WHB domain7 c2dsjA_
40.1
27
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside (thymidine) phosphorylase;PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
8 d1uoua1
38.9
21
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain9 c3iz5w_
37.3
17
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
10 d1o17a1
37.2
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain11 c3izcw_
35.7
20
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein rpl22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
12 c2fjrB_
28.6
7
PDB header: transcription regulatorChain: B: PDB Molecule: repressor protein ci;PDBTitle: crystal structure of bacteriophage 186
13 c2w1oA_
27.8
17
PDB header: translationChain: A: PDB Molecule: 60s acidic ribosomal protein p2;PDBTitle: nmr structure of dimerization domain of human ribosomal2 protein p2
14 c3h5qA_
26.6
11
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside phosphorylase;PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
15 d2f76x1
24.1
9
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: Mason-Pfizer monkey virus matrix protein16 d1n0ya_
23.8
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like17 c2k7bA_
23.6
8
PDB header: metal binding proteinChain: A: PDB Molecule: calcium-binding protein 1;PDBTitle: nmr structure of mg2+-bound cabp1 n-domain
18 d1sw8a_
23.0
9
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like19 d1khda1
21.2
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain20 d2elca1
19.4
11
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain21 c3ctnA_
not modelled
19.2
16
PDB header: calcium-binding proteinChain: A: PDB Molecule: troponin c;PDBTitle: structure of calcium-saturated cardiac troponin c, nmr, 302 structures
22 c1wlzD_
not modelled
18.5
13
PDB header: unknown functionChain: D: PDB Molecule: cap-binding protein complex interacting proteinPDBTitle: crystal structure of djbp fragment which was obtained by2 limited proteolysis
23 d1wlza1
not modelled
18.5
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins24 c1o17A_
not modelled
18.3
20
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
25 c2amiA_
not modelled
17.5
13
PDB header: cell cycleChain: A: PDB Molecule: caltractin;PDBTitle: solution structure of the calcium-loaded n-terminal sensor2 domain of centrin
26 c3evrA_
not modelled
17.0
14
PDB header: signaling proteinChain: A: PDB Molecule: myosin light chain kinase, green fluorescentPDBTitle: crystal structure of calcium bound monomeric gcamp2
27 d1sl7a_
not modelled
16.2
14
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like28 c2kdhA_
not modelled
15.5
16
PDB header: structural proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: the solution structure of human cardiac troponin c in2 complex with the green tea polyphenol; (-)-3 epigallocatechin-3-gallate
29 c2l98A_
not modelled
15.5
16
PDB header: contractile proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: structure of trans-resveratrol in complex with the cardiac regulatory2 protein troponin c
30 c3bjaA_
not modelled
15.2
9
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
31 c2igpA_
not modelled
14.9
14
PDB header: cell cycleChain: A: PDB Molecule: retinoblastoma-associated protein hec;PDBTitle: crystal structure of hec1 ch domain
32 d2pq3a1
not modelled
14.7
9
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like33 d1j55a_
not modelled
14.2
12
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins34 c2vqcA_
not modelled
13.9
42
PDB header: dna-binding proteinChain: A: PDB Molecule: hypothetical 13.2 kda protein;PDBTitle: structure of a dna binding winged-helix protein, f-112,2 from sulfolobus spindle-shaped virus 1.
35 d2vqca1
not modelled
13.9
42
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: F112-like36 d1dtla_
not modelled
13.5
14
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like37 d1xk4c1
not modelled
13.1
15
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins38 d1djxb1
not modelled
12.5
18
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins39 d2fcea1
not modelled
12.4
12
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like40 c2j0fC_
not modelled
11.9
24
PDB header: transferaseChain: C: PDB Molecule: thymidine phosphorylase;PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
41 c2g2bA_
not modelled
11.6
15
PDB header: immune systemChain: A: PDB Molecule: allograft inflammatory factor 1;PDBTitle: nmr structure of the human allograft inflammatory factor 1
42 d1wrka1
not modelled
11.5
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like43 d1fw4a_
not modelled
11.2
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like44 d1m39a_
not modelled
11.1
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like45 d1jc2a_
not modelled
11.1
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like46 c1jc2A_
not modelled
11.1
16
PDB header: structural proteinChain: A: PDB Molecule: troponin c, skeletal muscle;PDBTitle: complex of the c-domain of troponin c with residues 1-40 of2 troponin i
47 d1v8ga1
not modelled
10.7
16
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain48 c3cecA_
not modelled
10.6
18
PDB header: transcriptionChain: A: PDB Molecule: putative antidote protein of plasmid maintenance system;PDBTitle: crystal structure of a putative antidote protein of plasmid2 maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at3 1.60 a resolution
49 c3oiqB_
not modelled
10.3
27
PDB header: protein bindingChain: B: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of yeast telomere protein cdc13 ob1 and the2 catalytic subunit of dna polymerase alpha pol1
50 d1r2ua_
not modelled
10.1
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like51 d1vi0a2
not modelled
9.6
14
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain52 d1n0yb_
not modelled
9.3
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like53 c3pivA_
not modelled
9.2
17
PDB header: cytokineChain: A: PDB Molecule: interferon;PDBTitle: zebrafish interferon 1
54 d1eysh2
not modelled
9.2
47
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region55 c2fcdA_
not modelled
9.0
7
PDB header: cell cycleChain: A: PDB Molecule: myosin light chain 1;PDBTitle: solution structure of n-lobe myosin light chain from2 saccharomices cerevisiae
56 d1mzba_
not modelled
8.9
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like57 c2o8kA_
not modelled
8.8
12
PDB header: transcription/dnaChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: nmr structure of the sigma-54 rpon domain bound to the-242 promoter element
58 c1otpA_
not modelled
8.8
17
PDB header: phosphorylaseChain: A: PDB Molecule: thymidine phosphorylase;PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
59 c1irjG_
not modelled
8.5
12
PDB header: metal binding proteinChain: G: PDB Molecule: migration inhibitory factor-related protein 14;PDBTitle: crystal structure of the mrp14 complexed with chaps
60 c2b1uA_
not modelled
8.5
18
PDB header: metal binding proteinChain: A: PDB Molecule: calmodulin-like protein 5;PDBTitle: solution structure of calmodulin-like skin protein c2 terminal domain
61 c2k6oA_
not modelled
7.9
29
PDB header: antimicrobial proteinChain: A: PDB Molecule: cathelicidin antimicrobial peptide;PDBTitle: human ll-37 structure
62 c1brwB_
not modelled
7.9
15
PDB header: transferaseChain: B: PDB Molecule: protein (pyrimidine nucleoside phosphorylase);PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
63 d1f54a_
not modelled
7.9
7
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like64 d1avsa_
not modelled
7.8
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like65 c2d58A_
not modelled
7.8
13
PDB header: metal binding proteinChain: A: PDB Molecule: allograft inflammatory factor 1;PDBTitle: human microglia-specific protein iba1
66 c2jojA_
not modelled
7.8
11
PDB header: cell cycleChain: A: PDB Molecule: centrin protein;PDBTitle: nmr solution structure of n-terminal domain of euplotes2 octocarinatus centrin
67 c3jrtA_
not modelled
7.5
39
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: integron cassette protein vpc_cass2;PDBTitle: structure from the mobile metagenome of v. paracholerae:2 integron cassette protein vpc_cass2
68 c1ih0A_
not modelled
7.4
16
PDB header: contractile proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: structure of the c-domain of human cardiac troponin c in2 complex with ca2+ sensitizer emd 57033
69 d1ih0a_
not modelled
7.4
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like70 d2csfa1
not modelled
7.1
11
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain71 d1q5za_
not modelled
6.7
62
Fold: Invasion protein A (SipA) , C-terminal actin binding domainSuperfamily: Invasion protein A (SipA) , C-terminal actin binding domainFamily: Invasion protein A (SipA) , C-terminal actin binding domain72 c1q5zA_
not modelled
6.7
62
PDB header: cell invasionChain: A: PDB Molecule: sipa;PDBTitle: crystal structure of the c-terminal actin binding domain of2 salmonella invasion protein a (sipa)
73 d1cmga_
not modelled
6.7
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like74 c2aaoA_
not modelled
6.7
14
PDB header: transferaseChain: A: PDB Molecule: calcium-dependent protein kinase, isoform ak1;PDBTitle: regulatory apparatus of calcium dependent protein kinase from2 arabidopsis thaliana
75 c1sbjA_
not modelled
6.7
16
PDB header: contractile protein, structural proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: nmr structure of the mg2+-loaded c terminal domain of2 cardiac troponin c bound to the n terminal domain of3 cardiac troponin i
76 c1scvA_
not modelled
6.7
16
PDB header: contractile protein, structural proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i
77 c1fi5A_
not modelled
6.7
16
PDB header: contractile proteinChain: A: PDB Molecule: protein (troponin c);PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i.
78 d1fi5a_
not modelled
6.7
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like79 d1tn4a_
not modelled
6.6
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like80 c2k2aA_
not modelled
6.6
15
PDB header: contractile proteinChain: A: PDB Molecule: troponin c;PDBTitle: solution structure of the apo c terminal domain of lethocerus troponin2 c isoform f1
81 d1oqpa_
not modelled
6.5
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like82 c1ozsA_
not modelled
6.4
11
PDB header: structural proteinChain: A: PDB Molecule: troponin c, slow skeletal and cardiac muscles;PDBTitle: c-domain of human cardiac troponin c in complex with the2 inhibitory region of human cardiac troponin i
83 d1xrxa1
not modelled
6.4
67
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like84 c1xrxD_
not modelled
6.4
67
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
85 d1jw2a_
not modelled
6.3
19
Fold: Open three-helical up-and-down bundleSuperfamily: Hemolysin expression modulating protein HHAFamily: Hemolysin expression modulating protein HHA86 d2p5ka1
not modelled
6.1
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain87 d1tiza_
not modelled
6.1
12
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like88 c2e8mA_
not modelled
6.0
27
PDB header: signaling proteinChain: A: PDB Molecule: epidermal growth factor receptor kinasePDBTitle: solution structure of the c-terminal sam-domain of2 epidermal growth receptor pathway substrate 8
89 d1lj9a_
not modelled
5.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators90 d1a03a_
not modelled
5.8
10
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins91 c2a4jA_
not modelled
5.7
13
PDB header: structural proteinChain: A: PDB Molecule: centrin 2;PDBTitle: solution structure of the c-terminal domain (t94-y172) of2 the human centrin 2 in complex with a 17 residues peptide3 (p1-xpc) from xeroderma pigmentosum group c protein
92 d1jjcb2
not modelled
5.7
19
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Domains B1 and B5 of PheRS-beta, PheT93 c2ktgA_
not modelled
5.5
9
PDB header: ca-binding proteinChain: A: PDB Molecule: calmodulin, putative;PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein
94 d1b4aa1
not modelled
5.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain95 c2bcwC_
not modelled
5.4
20
PDB header: ribosomeChain: C: PDB Molecule: elongation factor g;PDBTitle: coordinates of the n-terminal domain of ribosomal protein2 l11,c-terminal domain of ribosomal protein l7/l12 and a3 portion of the g' domain of elongation factor g, as fitted4 into cryo-em map of an escherichia coli 70s*ef-5 g*gdp*fusidic acid complex
96 d1fi4a1
not modelled
5.4
22
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain97 d1f9na1
not modelled
5.3
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain98 d1np8a_
not modelled
5.3
12
Fold: EF Hand-likeSuperfamily: EF-handFamily: Penta-EF-hand proteins99 d1ap4a_
not modelled
5.2
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like