| 1 |
|
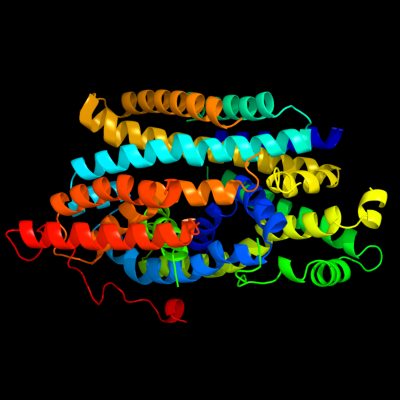
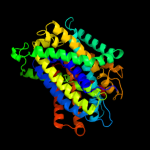
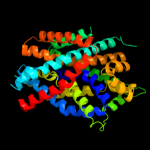
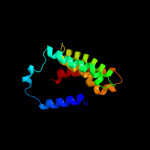
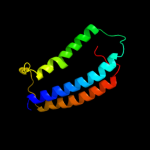
PDB 2xq2 chain A
Region: 4 - 498
Aligned: 473
Modelled: 473
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 2 |
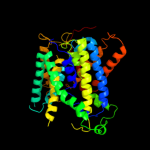
|
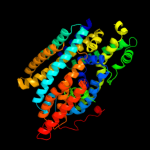
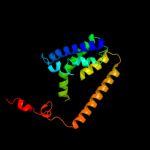
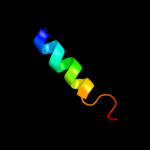
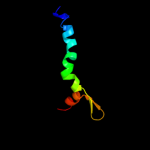
PDB 3dh4 chain A
Region: 26 - 497
Aligned: 465
Modelled: 453
Confidence: 100.0%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 3 |
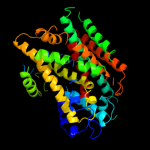
|
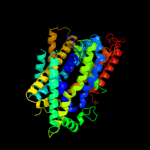
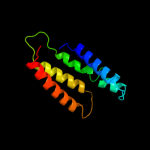
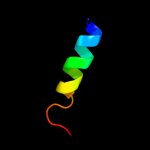
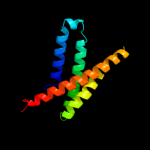
PDB 2jln chain A
Region: 28 - 490
Aligned: 435
Modelled: 435
Confidence: 99.7%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
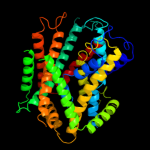
|
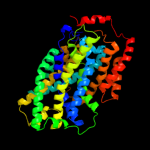
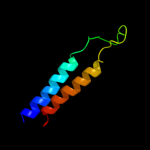
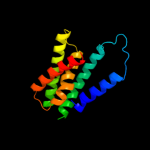
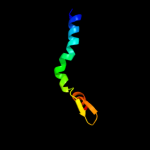
PDB 3gia chain A
Region: 39 - 472
Aligned: 413
Modelled: 423
Confidence: 99.7%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 5 |
|
PDB 3lrc chain C
Region: 39 - 478
Aligned: 396
Modelled: 396
Confidence: 99.3%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 6 |
|
PDB 2w8a chain C
Region: 6 - 421
Aligned: 402
Modelled: 384
Confidence: 98.2%
Identity: 10%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 7 |
|
PDB 3hfx chain A
Region: 2 - 421
Aligned: 406
Modelled: 406
Confidence: 97.6%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 8 |
|
PDB 2a65 chain A domain 1
Region: 43 - 424
Aligned: 373
Modelled: 373
Confidence: 96.8%
Identity: 12%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 9 |
|
PDB 3b9y chain A
Region: 337 - 501
Aligned: 161
Modelled: 164
Confidence: 29.7%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 10 |
|
PDB 3rlb chain A
Region: 373 - 478
Aligned: 104
Modelled: 106
Confidence: 22.4%
Identity: 20%
PDB header:thiamine-binding protein
Chain: A: PDB Molecule:thit;
PDBTitle: crystal structure at 2.0 a of the s-component for thiamin from an ecf-2 type abc transporter
Phyre2
| 11 |
|
PDB 3mk7 chain F
Region: 4 - 66
Aligned: 63
Modelled: 63
Confidence: 14.1%
Identity: 19%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 12 |
|
PDB 3hd6 chain A
Region: 340 - 474
Aligned: 135
Modelled: 135
Confidence: 8.7%
Identity: 10%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 13 |
|
PDB 2k9y chain B
Region: 12 - 30
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 26%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 14 |
|
PDB 2k9y chain A
Region: 12 - 30
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 26%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 15 |
|
PDB 1u7g chain A
Region: 340 - 475
Aligned: 135
Modelled: 132
Confidence: 7.7%
Identity: 16%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 16 |
|
PDB 4a2n chain B
Region: 1 - 106
Aligned: 106
Modelled: 106
Confidence: 6.4%
Identity: 9%
PDB header:transferase
Chain: B: PDB Molecule:isoprenylcysteine carboxyl methyltransferase;
PDBTitle: crystal structure of ma-icmt
Phyre2
| 17 |
|
PDB 3chx chain F
Region: 73 - 118
Aligned: 46
Modelled: 46
Confidence: 5.3%
Identity: 22%
PDB header:membrane protein
Chain: F: PDB Molecule:pmoa;
PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
Phyre2
| 18 |
|
PDB 3p5n chain A
Region: 372 - 483
Aligned: 110
Modelled: 112
Confidence: 5.3%
Identity: 21%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 19 |
|
PDB 1yew chain F
Region: 73 - 118
Aligned: 46
Modelled: 46
Confidence: 5.2%
Identity: 20%
PDB header:oxidoreductase, membrane protein
Chain: F: PDB Molecule:particulate methane monooxygenase, a subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 20 |
|
PDB 1a87 chain A
Region: 429 - 489
Aligned: 61
Modelled: 61
Confidence: 5.2%
Identity: 13%
PDB header:bacteriocin
Chain: A: PDB Molecule:colicin n;
PDBTitle: colicin n
Phyre2
| 21 |
|
| 22 |
|