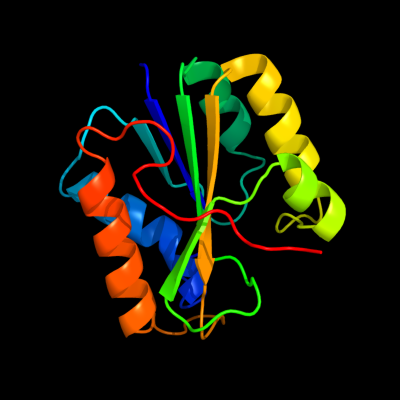
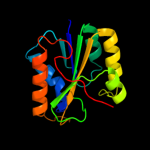
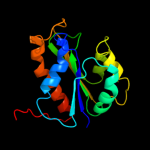
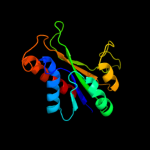
1 c2e85B_
100.0
99
PDB header: hydrolaseChain: B: PDB Molecule: hydrogenase 3 maturation protease;PDBTitle: crystal structure of the hydrogenase 3 maturation protease
2 d1cfza_
100.0
18
Fold: Phosphorylase/hydrolase-likeSuperfamily: HybD-likeFamily: Hydrogenase maturating endopeptidase HybD3 c3pu6A_
100.0
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of an uncharacterized protein from wolinella2 succinogenes
4 d1c8ba_
96.5
12
Fold: Phosphorylase/hydrolase-likeSuperfamily: HybD-likeFamily: Germination protease5 c3neaA_
94.4
19
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of peptidyl-trna hydrolase from francisella2 tularensis
6 d2ptha_
91.6
19
Fold: Phosphorylase/hydrolase-likeSuperfamily: Peptidyl-tRNA hydrolase-likeFamily: Peptidyl-tRNA hydrolase-like7 d1lssa_
82.2
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain8 c3l4bG_
81.9
11
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
9 c2z2jA_
81.7
16
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of peptidyl-trna hydrolase from mycobacterium2 tuberculosis
10 d2vapa1
78.4
13
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain11 c3eywA_
76.7
23
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
12 c1w5fA_
76.5
15
PDB header: cell divisionChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
13 d1w5fa1
76.1
15
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain14 c1w59B_
73.0
13
PDB header: cell divisionChain: B: PDB Molecule: cell division protein ftsz homolog 1;PDBTitle: ftsz dimer, empty (m. jannaschii)
15 c3rh0A_
71.5
27
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
16 c3vh3A_
71.1
24
PDB header: metal binding protein/protein transportChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of atg7ctd-atg8 complex
17 d2g2ca1
66.3
13
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like18 c2pjkA_
66.2
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 178aa long hypothetical molybdenum cofactorPDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
19 c3ic5A_
62.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
20 c3gucB_
60.4
23
PDB header: transferaseChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme 5;PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
21 c2zklA_
not modelled
60.4
14
PDB header: isomeraseChain: A: PDB Molecule: capsular polysaccharide synthesis enzyme cap5f;PDBTitle: crystal structure of capsular polysaccharide assembling protein capf2 from staphylococcus aureus
22 c3jviA_
not modelled
56.9
12
PDB header: hydrolaseChain: A: PDB Molecule: protein tyrosine phosphatase;PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
23 d1ryba_
not modelled
56.0
21
Fold: Phosphorylase/hydrolase-likeSuperfamily: Peptidyl-tRNA hydrolase-likeFamily: Peptidyl-tRNA hydrolase-like24 c2yxbA_
not modelled
55.2
11
PDB header: isomeraseChain: A: PDB Molecule: coenzyme b12-dependent mutase;PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
25 d2f1ka2
not modelled
51.9
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain26 c3ckyA_
not modelled
50.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-hydroxymethyl glutarate dehydrogenase;PDBTitle: structural and kinetic properties of a beta-hydroxyacid dehydrogenase2 involved in nicotinate fermentation
27 c3rfqC_
not modelled
49.8
17
PDB header: biosynthetic proteinChain: C: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase moab2;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
28 c3g17H_
not modelled
49.6
13
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: similar to 2-dehydropantoate 2-reductase;PDBTitle: structure of putative 2-dehydropantoate 2-reductase from2 staphylococcus aureus
29 c1u2pA_
not modelled
49.2
12
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
30 d1mv8a2
not modelled
49.1
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain31 c3gznB_
not modelled
49.0
26
PDB header: protein binding/ligaseChain: B: PDB Molecule: nedd8-activating enzyme e1 catalytic subunit;PDBTitle: structure of nedd8-activating enzyme in complex with nedd82 and mln4924
32 d1y1la_
not modelled
48.7
19
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases33 c3rofA_
not modelled
48.5
12
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-phosphatase ptpa;PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
34 c2z2vA_
not modelled
47.2
26
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
35 d1mkza_
not modelled
46.7
16
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like36 d1txga2
not modelled
45.4
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain37 c2vxyA_
not modelled
44.8
16
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: the structure of ftsz from bacillus subtilis at 1.7a2 resolution
38 c2ofpB_
not modelled
44.8
17
PDB header: oxidoreductaseChain: B: PDB Molecule: ketopantoate reductase;PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
39 c2f1kD_
not modelled
44.1
17
PDB header: oxidoreductaseChain: D: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
40 d2nqra3
not modelled
44.0
15
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like41 d1q0qa2
not modelled
43.8
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain42 d1ks9a2
not modelled
43.5
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain43 c3qhaB_
not modelled
43.4
33
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
44 c2fekA_
not modelled
43.2
20
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
45 c3c85A_
not modelled
42.9
21
PDB header: transport proteinChain: A: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
46 c2is8A_
not modelled
42.4
15
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
47 c2r6r1_
not modelled
42.2
13
PDB header: cell cycleChain: 1: PDB Molecule: cell division protein ftsz;PDBTitle: aquifex aeolicus ftsz
48 d1pgja2
not modelled
40.6
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain49 c2cwdA_
not modelled
40.6
14
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight phosphotyrosine protein phosphatase;PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
50 d1t2da1
not modelled
40.3
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like51 c1ks9A_
not modelled
40.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: ketopantoate reductase from escherichia coli
52 d1j5pa4
not modelled
39.3
40
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain53 d1n1ea2
not modelled
38.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain54 c2vawA_
not modelled
38.3
18
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz pseudomonas aeruginosa gdp
55 d1jlja_
not modelled
38.0
14
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like56 d1vl0a_
not modelled
37.9
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases57 c3vh1A_
not modelled
37.2
13
PDB header: metal binding proteinChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
58 c3lcmB_
not modelled
36.9
9
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
59 d1yovb1
not modelled
36.9
26
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)60 c3dojA_
not modelled
36.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase-like protein;PDBTitle: structure of glyoxylate reductase 1 from arabidopsis2 (atglyr1)
61 c1nhqA_
not modelled
35.9
16
PDB header: oxidoreductase (h2o2(a))Chain: A: PDB Molecule: nadh peroxidase;PDBTitle: crystallographic analyses of nadh peroxidase cys42ala and cys42ser2 mutants: active site structure, mechanistic implications, and an3 unusual environment of arg303
62 c2gf2B_
not modelled
35.6
27
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxyisobutyrate dehydrogenase;PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
63 c2rhoB_
not modelled
34.4
16
PDB header: cell cycleChain: B: PDB Molecule: cell division protein ftsz;PDBTitle: synthetic gene encoded bacillus subtilis ftsz ncs dimer with2 bound gdp and gtp-gamma-s
64 c1a5zA_
not modelled
33.8
23
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
65 c1vkzA_
not modelled
33.4
22
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: crystal structure of phosphoribosylamine--glycine ligase (tm1250) from2 thermotoga maritima at 2.30 a resolution
66 c1ojuA_
not modelled
33.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: 2.8 a resolution structure of malate dehydrogenase from2 archaeoglobus fulgidus in complex with etheno-nad.
67 d5pnta_
not modelled
33.1
14
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases68 c2cduB_
not modelled
33.0
13
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph oxidase;PDBTitle: the crystal structure of water-forming nad(p)h oxidase from2 lactobacillus sanfranciscensis
69 d1a5za1
not modelled
32.5
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like70 c2uyyD_
not modelled
31.8
13
PDB header: cytokineChain: D: PDB Molecule: n-pac protein;PDBTitle: structure of the cytokine-like nuclear factor n-pac
71 c2h1fB_
not modelled
31.6
16
PDB header: transferaseChain: B: PDB Molecule: lipopolysaccharide heptosyltransferase-1;PDBTitle: e. coli heptosyltransferase waac with adp
72 c2gi4A_
not modelled
31.5
12
PDB header: hydrolaseChain: A: PDB Molecule: possible phosphotyrosine protein phosphatase;PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
73 d1nhpa1
not modelled
30.9
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains74 d1ofua1
not modelled
30.4
18
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain75 c1zfnA_
not modelled
29.9
14
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase thif;PDBTitle: structural analysis of escherichia coli thif
76 c3fvwA_
not modelled
29.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative nad(p)h-dependent fmn reductase;PDBTitle: crystal structure of the q8dwd8_strmu protein from2 streptococcus mutans. northeast structural genomics3 consortium target smr99.
77 c1zggA_
not modelled
29.7
31
PDB header: hydrolaseChain: A: PDB Molecule: putative low molecular weight protein-tyrosine-PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
78 d1jw9b_
not modelled
28.5
11
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Molybdenum cofactor biosynthesis protein MoeB79 d1c1da1
not modelled
28.4
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain80 c3ghyA_
not modelled
28.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: ketopantoate reductase protein;PDBTitle: crystal structure of a putative ketopantoate reductase from ralstonia2 solanacearum molk2
81 d1yzfa1
not modelled
28.0
22
Fold: Flavodoxin-likeSuperfamily: SGNH hydrolaseFamily: TAP-like82 d1guza1
not modelled
27.6
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like83 d1rq2a1
not modelled
27.5
16
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain84 c3oc4A_
not modelled
27.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, pyridine nucleotide-disulfide family;PDBTitle: crystal structure of a pyridine nucleotide-disulfide family2 oxidoreductase from the enterococcus faecalis v583
85 c3qjaA_
not modelled
26.6
12
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
86 d1jf8a_
not modelled
26.5
19
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases87 c1ofuB_
not modelled
26.5
18
PDB header: bacterial cell division inhibitorChain: B: PDB Molecule: cell division protein ftsz;PDBTitle: crystal structure of sula:ftsz from pseudomonas aeruginosa
88 d1d1qa_
not modelled
25.9
15
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases89 d2blla1
not modelled
25.4
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases90 c2nvuB_
not modelled
25.3
27
PDB header: protein turnover, ligaseChain: B: PDB Molecule: maltose binding protein/nedd8-activating enzymePDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
91 d1uz5a3
not modelled
25.1
15
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like92 c3hp7A_
not modelled
24.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hemolysin, putative;PDBTitle: putative hemolysin from streptococcus thermophilus.
93 d2bfdb2
not modelled
24.7
21
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain94 d1ojua1
not modelled
24.6
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like95 c3lp8A_
not modelled
24.0
19
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine-glycine ligase;PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from2 ehrlichia chaffeensis
96 c3l6dB_
not modelled
23.9
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
97 c3grzA_
not modelled
23.9
21
PDB header: transferaseChain: A: PDB Molecule: ribosomal protein l11 methyltransferase;PDBTitle: crystal structure of ribosomal protein l11 methylase from2 lactobacillus delbrueckii subsp. bulgaricus
98 d1dg9a_
not modelled
23.8
18
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases99 c2dc1A_
not modelled
22.8
21
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
100 d1o0sa2
not modelled
22.7
21
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain101 d1y0ya2
not modelled
22.6
18
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases102 c2wmyH_
not modelled
22.5
14
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
103 c2g1uA_
not modelled
22.3
15
PDB header: transport proteinChain: A: PDB Molecule: hypothetical protein tm1088a;PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
104 c3prjB_
not modelled
21.6
16
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
105 d1a53a_
not modelled
21.5
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes106 d1i36a2
not modelled
21.3
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain107 d1v8da_
not modelled
21.2
14
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Hypothetical protein TT1679108 d1leha1
not modelled
21.1
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain109 d1a4ia1
not modelled
21.1
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain110 c2y0dB_
not modelled
21.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: bcec mutation y10k
111 c2qytA_
not modelled
21.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from porphyromonas2 gingivalis w83
112 d1jaya_
not modelled
20.4
37
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain113 c1d4fD_
not modelled
20.2
21
PDB header: hydrolaseChain: D: PDB Molecule: s-adenosylhomocysteine hydrolase;PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase