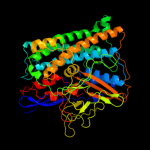
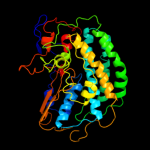
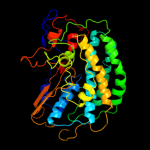
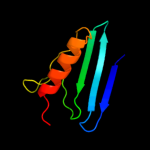
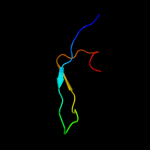
1 c3myrB_
100.0
50
PDB header: oxidoreductaseChain: B: PDB Molecule: nickel-dependent hydrogenase large subunit;PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
2 d1frfl_
100.0
45
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit3 d1wuil1
100.0
44
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit4 d1yq9h1
100.0
45
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit5 d1e3db_
100.0
44
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit6 c1h2aL_
100.0
45
PDB header: oxidoreductaseChain: L: PDB Molecule: hydrogenase;PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
7 d1cc1l_
100.0
36
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit8 c2wpnB_
100.0
39
PDB header: oxidoreductaseChain: B: PDB Molecule: periplasmic [nifese] hydrogenase, large subunit,PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
9 c2fug4_
100.0
21
PDB header: oxidoreductaseChain: 4: PDB Molecule: nadh-quinone oxidoreductase chain 4;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
10 d2fug41
100.0
21
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nqo4-like11 d1ffvc1
79.9
18
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like12 d1vqza1
74.9
7
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like13 c1vqzA_
73.3
6
PDB header: ligaseChain: A: PDB Molecule: lipoate-protein ligase, putative;PDBTitle: crystal structure of a putative lipoate-protein ligase a (sp_1160)2 from streptococcus pneumoniae tigr4 at 1.99 a resolution
14 d1x2ga1
71.1
13
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like15 d1t3qc1
57.8
15
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like16 d1yeya2
56.9
10
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like17 d1n62c1
52.1
15
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like18 c1xx3A_
51.4
25
PDB header: transport proteinChain: A: PDB Molecule: tonb protein;PDBTitle: solution structure of escherichia coli tonb-ctd
19 c2dgyA_
50.0
8
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
20 d2gskb1
49.6
25
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB21 d1o12a1
not modelled
48.2
33
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA22 d1u07a_
not modelled
45.4
25
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB23 d1jdfa2
not modelled
44.8
10
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like24 d1v97a4
not modelled
44.7
4
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like25 d1jroa3
not modelled
44.4
15
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like26 c2oqkA_
not modelled
43.6
14
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
27 d1iyxa2
not modelled
42.6
21
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like28 c2e5aA_
not modelled
41.8
6
PDB header: ligaseChain: A: PDB Molecule: lipoyltransferase 1;PDBTitle: crystal structure of bovine lipoyltransferase in complex2 with lipoyl-amp
29 d1rm6b1
not modelled
41.6
5
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like30 d1w6ta2
not modelled
41.5
26
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like31 c3o6uB_
not modelled
41.4
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein cpe2226;PDBTitle: crystal structure of cpe2226 protein from clostridium perfringens.2 northeast structural genomics consortium target cpr195
32 c2qq4A_
not modelled
40.9
21
PDB header: metal binding proteinChain: A: PDB Molecule: iron-sulfur cluster biosynthesis protein iscu;PDBTitle: crystal structure of iron-sulfur cluster biosynthesis2 protein iscu (ttha1736) from thermus thermophilus hb8
33 d1jt8a_
not modelled
40.0
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like34 c3mkcA_
not modelled
39.2
20
PDB header: isomeraseChain: A: PDB Molecule: racemase;PDBTitle: crystal structure of a putative racemase
35 c2grxC_
not modelled
38.9
25
PDB header: metal transportChain: C: PDB Molecule: protein tonb;PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
36 d1vk9a_
not modelled
38.8
27
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Hypothetical protein TM150637 c2dnwA_
not modelled
36.9
11
PDB header: transport proteinChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of rsgi ruh-059, an acp domain of acyl2 carrier protein, mitochondrial [precursor] from human cdna
38 d1su0b_
not modelled
36.5
13
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain39 d2ntka1
not modelled
36.3
23
Fold: Ntn hydrolase-likeSuperfamily: Archaeal IMP cyclohydrolase PurOFamily: Archaeal IMP cyclohydrolase PurO40 c1grjA_
not modelled
33.8
14
PDB header: transcription regulationChain: A: PDB Molecule: grea protein;PDBTitle: grea transcript cleavage factor from escherichia coli
41 d1d7qa_
not modelled
33.7
8
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like42 d3dm8a1
not modelled
32.2
23
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Rpa4348-like43 d1grja2
not modelled
31.3
16
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain44 d1e9ga_
not modelled
29.9
29
Fold: OB-foldSuperfamily: Inorganic pyrophosphataseFamily: Inorganic pyrophosphatase45 d1wfza_
not modelled
29.8
28
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain46 c1ufiD_
not modelled
29.8
30
PDB header: dna binding proteinChain: D: PDB Molecule: major centromere autoantigen b;PDBTitle: crystal structure of the dimerization domain of human cenp-b
47 d1ufia_
not modelled
29.6
30
Fold: ROP-likeSuperfamily: Dimerisation domain of CENP-BFamily: Dimerisation domain of CENP-B48 c2k9kA_
not modelled
28.1
19
PDB header: metal transportChain: A: PDB Molecule: tonb2;PDBTitle: molecular characterization of the tonb2 protein from vibrio2 anguillarum
49 d2f23a2
not modelled
27.9
24
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain50 d1xjsa_
not modelled
25.4
23
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain51 d1rvka2
not modelled
23.5
15
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like52 c2xzn5_
not modelled
23.3
57
PDB header: ribosomeChain: 5: PDB Molecule: ribosomal protein s26e containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
53 c3u5ga_
not modelled
23.3
57
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein s0-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
54 c3g0kA_
not modelled
22.9
20
PDB header: ca-binding proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: crystal structure of a protein of unknown function with a cystatin-2 like fold (saro_2880) from novosphingobium aromaticivorans dsm at3 1.30 a resolution
55 c1x2gB_
not modelled
21.6
15
PDB header: ligaseChain: B: PDB Molecule: lipoate-protein ligase a;PDBTitle: crystal structure of lipate-protein ligase a from2 escherichia coli
56 c1n62C_
not modelled
20.3
19
PDB header: oxidoreductaseChain: C: PDB Molecule: carbon monoxide dehydrogenase medium chain;PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
57 d1ihra_
not modelled
20.0
25
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB58 d1zjca1
not modelled
19.1
19
Fold: Thermophilic metalloprotease-likeSuperfamily: Thermophilic metalloprotease-likeFamily: Thermophilic metalloprotease (M29)59 c1y7jA_
not modelled
18.7
67
PDB header: signaling proteinChain: A: PDB Molecule: agouti signaling protein;PDBTitle: nmr structure family of human agouti signalling protein (80-2 132: q115y, s124y)
60 c3f40A_
not modelled
18.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized ntf2-like protein;PDBTitle: crystal structure of ntf2-like protein of unknown function2 (yp_677363.1) from cytophaga hutchinsonii atcc 33406 at3 1.27 a resolution
61 d2r4qa1
not modelled
18.5
24
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like62 d2q22a1
not modelled
18.2
32
Fold: Ava3019-likeSuperfamily: Ava3019-likeFamily: Ava3019-like63 c3b4oB_
not modelled
17.9
13
PDB header: biosynthetic proteinChain: B: PDB Molecule: phenazine biosynthesis protein a/b;PDBTitle: crystal structure of phenazine biosynthesis protein phza/b2 from burkholderia cepacia r18194, apo form
64 c2etnA_
not modelled
17.7
22
PDB header: transcriptionChain: A: PDB Molecule: anti-cleavage anti-grea transcription factorPDBTitle: crystal structure of thermus aquaticus gfh1
65 c3sc0A_
not modelled
17.5
75
PDB header: oxidoreductaseChain: A: PDB Molecule: methylmalonic aciduria and homocystinuria type c protein;PDBTitle: crystal structure of mmachc (1-238), a human b12 processing enzyme,2 complexed with methylcobalamin
66 d2nyga1
not modelled
17.3
30
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Aminoglycoside 3-N-acetyltransferase-like67 c3bmbB_
not modelled
16.0
18
PDB header: rna binding proteinChain: B: PDB Molecule: regulator of nucleoside diphosphate kinase;PDBTitle: crystal structure of a new rna polymerase interacting2 protein
68 d2auwa2
not modelled
15.8
11
Fold: NE0471 N-terminal domain-likeSuperfamily: NE0471 N-terminal domain-likeFamily: NE0471 N-terminal domain-like69 c3smaD_
not modelled
15.6
19
PDB header: transferaseChain: D: PDB Molecule: frbf;PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
70 d2chra2
not modelled
15.5
24
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like71 c2kzxA_
not modelled
15.4
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a3dht5 from clostridium thermocellum,2 northeast structural genomics consortium target cmr116
72 c2p4vA_
not modelled
15.4
17
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
73 c2w3rG_
not modelled
15.2
18
PDB header: oxidoreductaseChain: G: PDB Molecule: xanthine dehydrogenase;PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
74 c2fq2A_
not modelled
14.9
17
PDB header: lipid transportChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of minor conformation of holo-acyl2 carrier protein from malaria parasite plasmodium falciparum
75 c2nytB_
not modelled
14.4
67
PDB header: hydrolaseChain: B: PDB Molecule: probable c->u-editing enzyme apobec-2;PDBTitle: the apobec2 crystal structure and functional implications2 for aid
76 c3hk4B_
not modelled
13.9
18
PDB header: lyaseChain: B: PDB Molecule: mlr7391 protein;PDBTitle: crystal structure of a putative snoal-like polyketide cyclase2 [carbohydrate phosphatase] (mlr7391) from mesorhizobium loti at 1.963 a resolution
77 d2e1ba2
not modelled
13.8
31
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: ThrRS/AlaRS common domainFamily: AlaX-like78 d1ah9a_
not modelled
13.7
29
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 c2nqlB_
not modelled
13.6
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: isomerase/lactonizing enzyme;PDBTitle: crystal structure of a member of the enolase superfamily from2 agrobacterium tumefaciens
80 c3e4fB_
not modelled
13.6
21
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside n3-acetyltransferase;PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
81 c2dvyA_
not modelled
13.1
38
PDB header: hydrolaseChain: A: PDB Molecule: restriction endonuclease pabi;PDBTitle: crystal structure of restriction endonucleases pabi
82 c2auwB_
not modelled
13.1
11
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ne0471;PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
83 c3n6jA_
not modelled
12.9
27
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from actinobacillus succinogenes 130z
84 c3gkaB_
not modelled
12.9
17
PDB header: oxidoreductaseChain: B: PDB Molecule: n-ethylmaleimide reductase;PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
85 d1gsma2
not modelled
12.9
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains86 c2l9fA_
not modelled
12.5
12
PDB header: transferaseChain: A: PDB Molecule: cale8;PDBTitle: nmr solution structure of meacp
87 c2kboA_
not modelled
12.4
56
PDB header: hydrolaseChain: A: PDB Molecule: dna dc->du-editing enzyme apobec-3g;PDBTitle: structure, interaction, and real-time monitoring of the2 enzymatic reaction of wild type apobec3g
88 c3f14A_
not modelled
12.1
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized ntf2-like protein;PDBTitle: crystal structure of ntf2-like protein of unknown function2 (yp_680363.1) from cytophaga hutchinsonii atcc 33406 at 1.45 a3 resolution
89 c2l4bA_
not modelled
12.1
8
PDB header: transferaseChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of a putative acyl carrier protein from anaplasma2 phagocytophilum. seattle structural genomics center for infectious3 disease target anpha.01018.a
90 d1h9aa2
not modelled
12.1
16
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Glucose 6-phosphate dehydrogenase-like91 d1t8ka_
not modelled
12.0
19
Fold: Acyl carrier protein-likeSuperfamily: ACP-likeFamily: Acyl-carrier protein (ACP)92 c3bjsB_
not modelled
11.5
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of a member of enolase superfamily from polaromonas2 sp. js666
93 c1t3qF_
not modelled
11.4
16
PDB header: oxidoreductaseChain: F: PDB Molecule: quinoline 2-oxidoreductase medium subunit;PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
94 d2etna2
not modelled
11.2
22
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain95 c2kciA_
not modelled
11.1
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative acyl carrier protein;PDBTitle: solution nmr structure of gmet_2339 from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr141
96 d1z1sa1
not modelled
11.1
25
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: PhzA/PhzB-like97 c3nxlD_
not modelled
10.9
15
PDB header: lyaseChain: D: PDB Molecule: glucarate dehydratase;PDBTitle: crystal structure of glucarate dehydratase from burkholderia cepacia2 complexed with magnesium
98 c2pn0D_
not modelled
10.7
14
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
99 c3fgyB_
not modelled
10.6
0
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized ntf2-like protein;PDBTitle: crystal structure of a ntf2-like protein (bxe_b1094) from burkholderia2 xenovorans lb400 at 1.59 a resolution