| 1 | c1mbbA_ |
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uridine diphospho-n-acetylenolpyruvylglucosamine
PDBTitle: oxidoreductase
|
| 2 | c3i99A_ |
|
 |
100.0 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: the crystal structure of the udp-n-acetylenolpyruvoylglucosamine2 reductase from the vibrio cholerae o1 biovar tor
|
| 3 | c1hskA_ |
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of s. aureus murb
|
| 4 | c2gquA_ |
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvylglucosamine reductase;
PDBTitle: crystal structure of udp-n-acetylenolpyruvylglucosamine2 reductase (murb) from thermus caldophilus
|
| 5 | d1uxya2 |
|
 |
100.0 |
99 |
Fold:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Superfamily:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain |
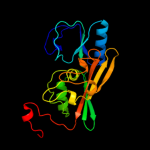
| 6 | d1uxya1 |
|
 |
100.0 |
100 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
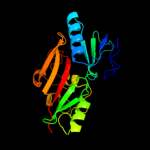
| 7 | d1hska1 |
|
 |
100.0 |
25 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
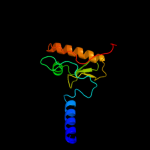
| 8 | d1hska2 |
|
 |
100.0 |
36 |
Fold:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Superfamily:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain |
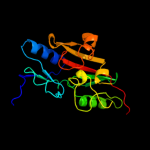
| 9 | c3fwaA_ |
|
 |
99.9 |
11 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
| 10 | c2uuvC_ |
|
 |
99.9 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
| 11 | c3d2hA_ |
|
 |
99.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:berberine bridge-forming enzyme;
PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
|
| 12 | c3bw7A_ |
|
 |
99.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 1;
PDBTitle: maize cytokinin oxidase/dehydrogenase complexed with the allenic2 cytokinin analog ha-1
|
| 13 | c2bvfA_ |
|
 |
99.9 |
14 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
| 14 | c3pm9A_ |
|
 |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
| 15 | c1wveB_ |
|
 |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
| 16 | c1zr6A_ |
|
 |
99.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
| 17 | c1i19B_ |
|
 |
99.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
| 18 | c3popD_ |
|
 |
99.9 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
| 19 | c2exrA_ |
|
 |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 7;
PDBTitle: x-ray structure of cytokinin oxidase/dehydrogenase (ckx)2 from arabidopsis thaliana at5g21482
|
| 20 | c2wdwB_ |
|
 |
99.9 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
| 21 | c2y3rC_ |
|
not modelled |
99.9 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent2 tirandamycin oxidase taml in p21 space group
|
| 22 | c1ahuB_ |
|
not modelled |
99.9 |
18 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
| 23 | c2vfvA_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xylitol oxidase;
PDBTitle: alditol oxidase from streptomyces coelicolor a3(2): complex2 with sulphite
|
| 24 | c2ipiD_ |
|
not modelled |
99.9 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
| 25 | d1wvfa2 |
|
not modelled |
99.9 |
16 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 26 | c1f0xA_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral2 membrane respiratory enzyme.
|
| 27 | c3js8A_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: solvent-stable cholesterol oxidase
|
| 28 | d1e8ga2 |
|
not modelled |
99.9 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 29 | d2i0ka2 |
|
not modelled |
99.9 |
16 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 30 | d1w1oa2 |
|
not modelled |
99.9 |
12 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 31 | d1f0xa2 |
|
not modelled |
99.9 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 32 | c2yvsA_ |
|
not modelled |
99.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycolate oxidase subunit glce;
PDBTitle: crystal structure of glycolate oxidase subunit glce from thermus2 thermophilus hb8
|
| 33 | d1n62c2 |
|
not modelled |
98.4 |
14 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 34 | d1ffvc2 |
|
not modelled |
98.3 |
15 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 35 | c1n62C_ |
|
not modelled |
98.3 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbon monoxide dehydrogenase medium chain;
PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
|
| 36 | d3b9jb2 |
|
not modelled |
98.2 |
10 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 37 | c3hrdC_ |
|
not modelled |
98.2 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinate dehydrogenase fad-subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 38 | c1ffuF_ |
|
not modelled |
98.1 |
14 |
PDB header:hydrolase
Chain: F: PDB Molecule:cutm, flavoprotein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 39 | d1v97a6 |
|
not modelled |
98.1 |
11 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 40 | c1t3qF_ |
|
not modelled |
98.1 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:quinoline 2-oxidoreductase medium subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 41 | c3etrM_ |
|
not modelled |
98.0 |
12 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of xanthine oxidase in complex with2 lumazine
|
| 42 | c3b9jJ_ |
|
not modelled |
98.0 |
12 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:xanthine oxidase;
PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
|
| 43 | d1rm6b2 |
|
not modelled |
97.9 |
19 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 44 | d1t3qc2 |
|
not modelled |
97.9 |
19 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 45 | c1wygA_ |
|
not modelled |
97.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
|
| 46 | c2w3rG_ |
|
not modelled |
97.7 |
14 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
| 47 | c1rm6E_ |
|
not modelled |
97.5 |
19 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 48 | d1jroa4 |
|
not modelled |
97.4 |
10 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 49 | c1xtyD_ |
|
not modelled |
65.8 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna2 hydrolase
|
| 50 | d1q7sa_ |
|
not modelled |
64.1 |
21 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 51 | d1rlka_ |
|
not modelled |
60.6 |
15 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 52 | c2d3kA_ |
|
not modelled |
55.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
|
| 53 | c2zv3E_ |
|
not modelled |
54.3 |
21 |
PDB header:hydrolase
Chain: E: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
|
| 54 | c2r47C_ |
|
not modelled |
53.9 |
24 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein mth_862;
PDBTitle: crystal structure of mth_862 protein of unknown function from2 methanothermobacter thermautotrophicus
|
| 55 | c2l1nA_ |
|
not modelled |
52.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the protein yp_399305.1
|
| 56 | d3erja1 |
|
not modelled |
48.3 |
10 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 57 | c2qiwA_ |
|
not modelled |
40.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 58 | c3ih1A_ |
|
not modelled |
39.1 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 59 | c2ze3A_ |
|
not modelled |
32.5 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 60 | d1ik6a1 |
|
not modelled |
31.6 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 61 | c2yswB_ |
|
not modelled |
30.5 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of the 3-dehydroquinate dehydratase from aquifex2 aeolicus vf5
|
| 62 | c3dfeA_ |
|
not modelled |
27.3 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:putative pii-like signaling protein;
PDBTitle: crystal structure of a putative pii-like signaling protein2 (yp_323533.1) from anabaena variabilis atcc 29413 at 2.35 a3 resolution
|
| 63 | c1ik6A_ |
|
not modelled |
26.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate dehydrogenase;
PDBTitle: 3d structure of the e1beta subunit of pyruvate2 dehydrogenase from the archeon pyrobaculum aerophilum
|
| 64 | c1bplB_ |
|
not modelled |
26.5 |
20 |
PDB header:glycosyltransferase
Chain: B: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
| 65 | d1jaka1 |
|
not modelled |
24.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 66 | c2ylaA_ |
|
not modelled |
24.3 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 67 | d1hx0a2 |
|
not modelled |
23.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 68 | c2hjpA_ |
|
not modelled |
23.6 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 69 | c2yl8A_ |
|
not modelled |
23.2 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 70 | d1ybha1 |
|
not modelled |
23.2 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 71 | d1w85b1 |
|
not modelled |
22.7 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 72 | d1umdb1 |
|
not modelled |
22.4 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 73 | c3mt1B_ |
|
not modelled |
22.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:putative carboxynorspermidine decarboxylase protein;
PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
|
| 74 | d1yhta1 |
|
not modelled |
22.2 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 75 | d2bdua1 |
|
not modelled |
21.2 |
29 |
Fold:HAD-like
Superfamily:HAD-like
Family:Pyrimidine 5'-nucleotidase (UMPH-1) |
| 76 | d2ozlb1 |
|
not modelled |
21.1 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 77 | c2grvC_ |
|
not modelled |
21.0 |
10 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:lpqw;
PDBTitle: crystal structure of lpqw
|
| 78 | d2pgca1 |
|
not modelled |
20.6 |
36 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Marine metagenome family DABB3 |
| 79 | c1bplA_ |
|
not modelled |
20.3 |
14 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
| 80 | d1ne9a1 |
|
not modelled |
19.6 |
19 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:FemXAB nonribosomal peptidyltransferases |
| 81 | d2bfdb1 |
|
not modelled |
19.6 |
11 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 82 | d1jaea2 |
|
not modelled |
19.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 83 | d1nowa1 |
|
not modelled |
18.9 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 84 | c3gh7A_ |
|
not modelled |
18.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of beta-hexosaminidase from paenibacillus2 sp. ts12 in complex with galnac
|
| 85 | d3dhpa2 |
|
not modelled |
18.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 86 | d2gjxa1 |
|
not modelled |
18.6 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 87 | d1qs0b1 |
|
not modelled |
18.1 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 88 | c1zlpA_ |
|
not modelled |
18.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 89 | c3nsnA_ |
|
not modelled |
17.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylglucosaminidase;
PDBTitle: crystal structure of insect beta-n-acetyl-d-hexosaminidase ofhex12 complexed with tmg-chitotriomycin
|
| 90 | c1hvxA_ |
|
not modelled |
17.6 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: bacillus stearothermophilus alpha-amylase
|
| 91 | c1nouA_ |
|
not modelled |
17.5 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase beta chain;
PDBTitle: native human lysosomal beta-hexosaminidase isoform b
|
| 92 | c2gjxE_ |
|
not modelled |
17.4 |
29 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta-hexosaminidase alpha chain;
PDBTitle: crystallographic structure of human beta-hexosaminidase a
|
| 93 | d1gcya2 |
|
not modelled |
17.1 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 94 | d1qbaa3 |
|
not modelled |
17.1 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 95 | c1m8oB_ |
|
not modelled |
16.9 |
17 |
PDB header:membrane protein
Chain: B: PDB Molecule:platele integrin beta3 subunit: cytoplasmic
PDBTitle: platelet integrin alfaiib-beta3 cytoplasmic domain
|
| 96 | d1ps9a1 |
|
not modelled |
16.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 97 | c3rcnA_ |
|
not modelled |
16.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of beta-n-acetylhexosaminidase from arthrobacter2 aurescens
|
| 98 | c1m04A_ |
|
not modelled |
16.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: mutant streptomyces plicatus beta-hexosaminidase (d313n) in complex2 with product (glcnac)
|
| 99 | c2g0cA_ |
|
not modelled |
16.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent rna helicase dbpa;
PDBTitle: structure of the rna binding domain (residues 404-479) of the bacillus2 subtilis yxin protein
|

