| 1 |
|
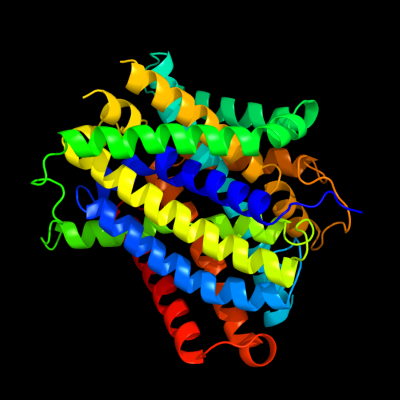
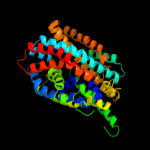
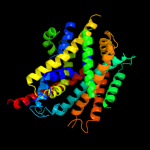
PDB 3gia chain A
Region: 7 - 407
Aligned: 395
Modelled: 395
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
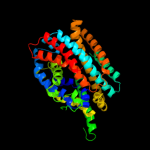
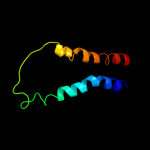
PDB 3lrc chain C
Region: 7 - 398
Aligned: 373
Modelled: 373
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
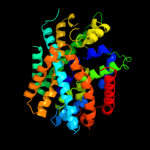
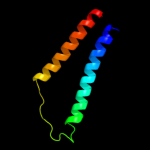
PDB 2jln chain A
Region: 2 - 407
Aligned: 399
Modelled: 406
Confidence: 99.9%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
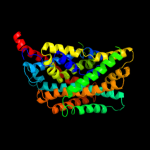
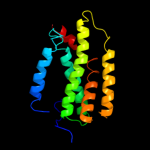
PDB 3dh4 chain A
Region: 9 - 404
Aligned: 385
Modelled: 385
Confidence: 98.5%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 5 |
|
PDB 2xq2 chain A
Region: 9 - 404
Aligned: 385
Modelled: 385
Confidence: 98.5%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 12 - 411
Aligned: 397
Modelled: 397
Confidence: 97.9%
Identity: 12%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 4 - 380
Aligned: 374
Modelled: 377
Confidence: 96.5%
Identity: 10%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 6 - 376
Aligned: 369
Modelled: 369
Confidence: 75.3%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 1fft chain B domain 2
Region: 343 - 410
Aligned: 68
Modelled: 68
Confidence: 8.5%
Identity: 3%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 10 |
|
PDB 2hg5 chain D
Region: 12 - 42
Aligned: 31
Modelled: 31
Confidence: 7.6%
Identity: 26%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 11 |
|
PDB 3dtu chain B domain 2
Region: 342 - 410
Aligned: 69
Modelled: 69
Confidence: 6.9%
Identity: 12%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 1xg8 chain A
Region: 66 - 86
Aligned: 21
Modelled: 21
Confidence: 6.8%
Identity: 14%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: YuzD-like
Phyre2
| 13 |
|
PDB 1iwg chain A domain 8
Region: 215 - 376
Aligned: 162
Modelled: 162
Confidence: 6.8%
Identity: 10%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 14 |
|
PDB 1qle chain B
Region: 342 - 410
Aligned: 69
Modelled: 69
Confidence: 6.6%
Identity: 14%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2
| 15 |
|
PDB 1ar1 chain B
Region: 342 - 410
Aligned: 69
Modelled: 69
Confidence: 6.6%
Identity: 14%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 16 |
|
PDB 2p9x chain B
Region: 64 - 96
Aligned: 31
Modelled: 33
Confidence: 6.5%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph0832;
PDBTitle: crystal structure of ph0832 from pyrococcus horikoshii ot3
Phyre2
| 17 |
|
PDB 2knc chain B
Region: 8 - 32
Aligned: 25
Modelled: 25
Confidence: 5.8%
Identity: 24%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 18 |
|
PDB 1nek chain D
Region: 145 - 248
Aligned: 103
Modelled: 104
Confidence: 5.2%
Identity: 12%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 19 |
|
PDB 3ehb chain B domain 2
Region: 342 - 410
Aligned: 69
Modelled: 69
Confidence: 5.2%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2