1 c1zeqX_
100.0
99
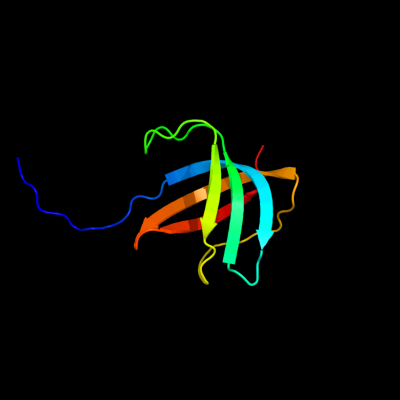
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
2 c2l55A_
99.9
27
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
3 c3camB_
95.2
20
PDB header: gene regulationChain: B: PDB Molecule: cold-shock domain family protein;PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
4 c2k5nA_
94.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cold-shock protein;PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
5 c3trzE_
91.0
15
PDB header: rna binding protein/rnaChain: E: PDB Molecule: protein lin-28 homolog a;PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
6 d2es2a1
90.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like7 c2kcmA_
87.9
18
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: cold shock domain family protein;PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
8 d1mjca_
86.2
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like9 d1c9oa_
84.2
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like10 c3aqqD_
82.8
13
PDB header: dna binding proteinChain: D: PDB Molecule: calcium-regulated heat stable protein 1;PDBTitle: crystal structure of human crhsp-24
11 c2ytxA_
81.6
18
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the second cold-shock domain of the human2 kiaa0885 protein (unr protein)
12 d1g6pa_
76.4
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like13 d1h95a_
76.2
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like14 c3tdqB_
75.0
21
PDB header: cell adhesionChain: B: PDB Molecule: pily2 protein;PDBTitle: crystal structure of a fimbrial biogenesis protein pily22 (pily2_pa4555) from pseudomonas aeruginosa pao1 at 2.10 a resolution
15 c2ytvA_
72.6
15
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fifth cold-shock domain of the human2 kiaa0885 protein (unr protein)
16 c1x65A_
71.6
14
PDB header: rna binding proteinChain: A: PDB Molecule: unr protein;PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
17 d1wfqa_
68.3
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like18 c3d0fA_
68.1
8
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
19 d2f5va2
62.8
23
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GMC oxidoreductases20 c2vc8A_
61.9
28
PDB header: protein-bindingChain: A: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of the lsm domain of human edc3 (enhancer2 of decapping 3)
21 c3mmlE_
not modelled
60.2
10
PDB header: hydrolaseChain: E: PDB Molecule: allophanate hydrolase subunit 2;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
22 c3a0jB_
not modelled
57.8
20
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
23 c2khiA_
not modelled
56.0
14
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
24 d1wida_
not modelled
55.2
21
Fold: DNA-binding pseudobarrel domainSuperfamily: DNA-binding pseudobarrel domainFamily: B3 DNA binding domain25 d1qbaa2
not modelled
48.9
23
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Bacterial chitobiase, n-terminal domain26 c3kf6B_
not modelled
47.6
29
PDB header: structural proteinChain: B: PDB Molecule: protein ten1;PDBTitle: crystal structure of s. pombe stn1-ten1 complex
27 d1x9la_
not modelled
43.4
17
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: DR1885-like metal-binding proteinFamily: DR1885-like metal-binding protein28 d2ba0a1
not modelled
42.5
12
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like29 d1hh2p1
not modelled
39.8
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like30 c3u7zA_
not modelled
39.4
14
PDB header: metal binding proteinChain: A: PDB Molecule: putative metal binding protein rumgna_00854;PDBTitle: crystal structure of a putative metal binding protein rumgna_008542 (zp_02040092.1) from ruminococcus gnavus atcc 29149 at 1.30 a3 resolution
31 c2vzaD_
not modelled
38.9
31
PDB header: cell adhesionChain: D: PDB Molecule: cell filamentation protein;PDBTitle: type iv secretion system effector protein bepa
32 c2ytyA_
not modelled
38.1
13
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fourth cold-shock domain of the human2 kiaa0885 protein (unr protein)
33 c3m7aA_
not modelled
37.7
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of saro_0823 (yp_496102.1) a protein of2 unknown function from novosphingobium aromaticivorans dsm3 12444 at 1.22 a resolution
34 c3ayhB_
not modelled
36.6
19
PDB header: transcriptionChain: B: PDB Molecule: dna-directed rna polymerase iii subunit rpc8;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
35 d1wi5a_
not modelled
29.4
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like36 d1gvha2
not modelled
29.2
8
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like37 d1gyca1
not modelled
28.2
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins38 c2k6zA_
not modelled
28.1
21
PDB header: metal transportChain: A: PDB Molecule: putative uncharacterized protein ttha1943;PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
39 c3i18A_
not modelled
27.7
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2051 protein;PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
40 d2phcb1
not modelled
27.4
22
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like41 c2phcB_
not modelled
27.3
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
42 d2f23a2
not modelled
25.6
10
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain43 c2h47C_
not modelled
25.6
29
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: azurin;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
44 d1xnea_
not modelled
25.5
24
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain45 d1qnia1
not modelled
25.2
25
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Nitrosocyanin46 d1u0la1
not modelled
25.0
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like47 c1t9hA_
not modelled
24.1
21
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: the crystal structure of yloq, a circularly permuted gtpase.
48 d2f1la1
not modelled
23.9
35
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: RimM C-terminal domain-like49 c2rf4A_
not modelled
23.5
15
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase i subunit rpa4;PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
50 c3mmlD_
not modelled
23.4
30
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
51 c3ie5A_
not modelled
22.3
11
PDB header: plant protein, biosynthetic proteinChain: A: PDB Molecule: phenolic oxidative coupling protein hyp-1;PDBTitle: crystal structure of hyp-1 protein from hypericum perforatum2 (st john's wort) involved in hypericin biosynthesis
52 d2cnda1
not modelled
21.5
11
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like53 d1v6ga2
not modelled
21.4
42
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain54 d1fwxa1
not modelled
20.8
25
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Nitrosocyanin55 c2aanA_
not modelled
20.0
19
PDB header: electron transportChain: A: PDB Molecule: auracyanin a;PDBTitle: auracyanin a: a "blue" copper protein from the green thermophilic2 photosynthetic bacterium,chloroflexus aurantiacus
56 c2khjA_
not modelled
19.7
15
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
57 d1tyeb1
not modelled
19.6
31
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Integrin domainsFamily: Integrin domains58 d1cqxa2
not modelled
19.1
3
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like59 c2zp2B_
not modelled
19.1
44
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
60 d1joia_
not modelled
18.8
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like61 d1t9ha1
not modelled
18.7
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like62 c2p4vA_
not modelled
18.3
13
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
63 d2ccwa1
not modelled
18.2
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like64 d2c35b1
not modelled
17.8
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like65 d1ndha1
not modelled
17.4
18
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like66 c3oepA_
not modelled
17.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha0988;PDBTitle: crystal structure of ttha0988 in space group p43212
67 d1cc3a_
not modelled
17.1
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like68 d1i94q_
not modelled
16.9
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like69 d1a5ra_
not modelled
16.3
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related70 d2etna2
not modelled
16.1
10
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain71 d2z0sa1
not modelled
16.0
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like72 d1ws8a_
not modelled
15.7
31
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like73 c2kjpA_
not modelled
15.4
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ylbl;PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
74 d1t2wa_
not modelled
15.3
20
Fold: SortaseSuperfamily: SortaseFamily: Sortase75 c3pjyB_
not modelled
15.0
15
PDB header: transcription regulatorChain: B: PDB Molecule: hypothetical signal peptide protein;PDBTitle: crystal structure of a putative transcription regulator (r01717) from2 sinorhizobium meliloti 1021 at 1.55 a resolution
76 d1k0ha_
not modelled
15.0
6
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: gpFII-like77 d1azca_
not modelled
14.9
14
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like78 d1q46a2
not modelled
14.9
10
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 d1gawa1
not modelled
14.8
15
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like80 c2w1kB_
not modelled
14.7
17
PDB header: transferaseChain: B: PDB Molecule: putative sortase;PDBTitle: crystal structure of sortase c-3 (srtc-3) from2 streptococcus pneumoniae
81 d2z9ia1
not modelled
14.5
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases82 d1jb9a1
not modelled
14.5
28
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like83 d1nppa2
not modelled
14.1
35
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain84 d1nz9a_
not modelled
14.0
35
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain85 c3qo6B_
not modelled
13.9
17
PDB header: photosynthesisChain: B: PDB Molecule: protease do-like 1, chloroplastic;PDBTitle: crystal structure analysis of the plant protease deg1
86 d1qfza1
not modelled
13.6
19
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like87 d2gnra1
not modelled
13.3
10
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: SSO2064-like88 c3mjjD_
not modelled
13.3
20
PDB header: hydrolaseChain: D: PDB Molecule: predicted acetamidase/formamidase;PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
89 d2i6va1
not modelled
13.2
8
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like90 d1n67a1
not modelled
13.0
19
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Fibrinogen-binding domain91 c2l66B_
not modelled
12.9
32
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
92 d1jzga_
not modelled
12.6
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like93 c2rcnA_
not modelled
12.5
15
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
94 d1t3qb1
not modelled
12.2
12
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like95 c3ke2A_
not modelled
12.2
31
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yp_928783.1;PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
96 c3fp9E_
not modelled
12.1
29
PDB header: hydrolaseChain: E: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
97 d1plaa_
not modelled
12.1
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like98 d1v10a2
not modelled
11.8
14
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins99 c2kw8A_
not modelled
11.8
13
PDB header: protein bindingChain: A: PDB Molecule: lpxtg-site transpeptidase family protein;PDBTitle: solution structure of bacillus anthracis sortase a (srta)2 transpeptidase