1 c1a6qA_
100.0
16
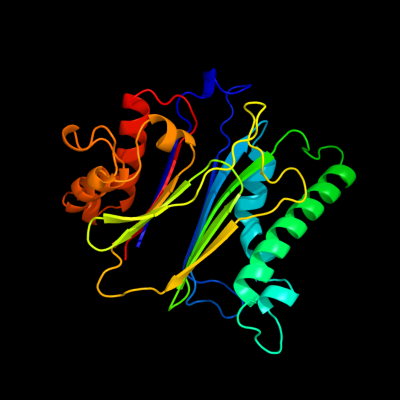
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase 2c;PDBTitle: crystal structure of the protein serine/threonine phosphatase 2c at 22 a resolution
2 d1txoa_
100.0
16
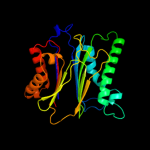
Fold: PP2C-likeSuperfamily: PP2C-likeFamily: PP2C-like3 c2pk0C_
100.0
15
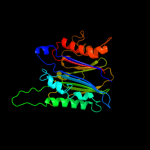
PDB header: signaling proteinChain: C: PDB Molecule: serine/threonine protein phosphatase stp1;PDBTitle: structure of the s. agalactiae serine/threonine phosphatase at 2.652 resolution
4 d1a6qa2
100.0
16
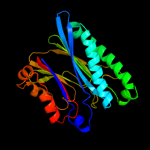
Fold: PP2C-likeSuperfamily: PP2C-likeFamily: PP2C-like5 c3kdjB_
100.0
15
PDB header: hydrolase/hormone receptorChain: B: PDB Molecule: protein phosphatase 2c 56;PDBTitle: complex structure of (+)-aba-bound pyl1 and abi1
6 c2i44A_
100.0
13
PDB header: hydrolaseChain: A: PDB Molecule: serine-threonine phosphatase 2c;PDBTitle: crystal structure of serine-threonine phosphatase 2c from2 toxoplasma gondii
7 c2cm1A_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: serine threonine protein phosphatase pstp;PDBTitle: crystal structure of the catalytic domain of serine2 threonine protein phosphatase pstp in complex with3 2 manganese ions.
8 c3kb3B_
100.0
15
PDB header: signaling proteinChain: B: PDB Molecule: protein phosphatase 2c 16;PDBTitle: crystal structure of abscisic acid-bound pyl2 in complex with hab1
9 c2jfsA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: ser-thr phosphatase mspp;PDBTitle: crystal structure of the ppm ser-thr phosphatase mspp from2 mycobacterium smegmatis in complex with cacodylate
10 c2i0oA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: ser/thr phosphatase;PDBTitle: crystal structure of anopheles gambiae ser/thr phosphatase complexed2 with zn2+
11 c2iq1A_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: protein phosphatase 2c kappa, ppm1k;PDBTitle: crystal structure of human ppm1k
12 c2irmA_
100.0
11
PDB header: transferaseChain: A: PDB Molecule: mitogen-activated protein kinase kinase kinase 7PDBTitle: crystal structure of mitogen-activated protein kinase kinase kinase 72 interacting protein 1 from anopheles gambiae
13 c2isnB_
100.0
14
PDB header: hydrolaseChain: B: PDB Molecule: nysgxrc-8828z, phosphatase;PDBTitle: crystal structure of a phosphatase from a pathogenic strain toxoplasma2 gondii
14 c2pnqA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: [pyruvate dehydrogenase [lipoamide]]-phosphatasePDBTitle: crystal structure of pyruvate dehydrogenase phosphatase 12 (pdp1)
15 c2j82A_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: protein serine-threonine phosphatase;PDBTitle: structural analysis of the pp2c family phosphatase tppha2 from thermosynechococcus elongatus
16 c2pomA_
99.9
12
PDB header: signaling protein/metal binding proteinChain: A: PDB Molecule: mitogen-activated protein kinase kinase kinase 7-PDBTitle: tab1 with manganese ion
17 c2j4oA_
99.9
12
PDB header: protein bindingChain: A: PDB Molecule: mitogen-activated protein kinase kinase kinasePDBTitle: structure of tab1
18 c3rnrB_
99.9
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: stage ii sporulation e family protein;PDBTitle: crystal structure of stage ii sporulation e family protein from2 thermanaerovibrio acidaminovorans
19 c3d8kD_
99.9
14
PDB header: hydrolaseChain: D: PDB Molecule: protein phosphatase 2c;PDBTitle: crsytal structure of a phosphatase from a toxoplasma gondii
20 c3pu9A_
99.8
14
PDB header: transferaseChain: A: PDB Molecule: protein serine/threonine phosphatase;PDBTitle: crystal structure of serine/threonine phosphatase sphaerobacter2 thermophilus dsm 20745
21 c3t9qB_
not modelled
99.6
11
PDB header: hydrolaseChain: B: PDB Molecule: stage ii sporulation protein e;PDBTitle: structure of the phosphatase domain of the cell fate determinant2 spoiie from bacillus subtilis (mn presoaked)
22 c3ke6A_
not modelled
98.8
12
PDB header: unknown functionChain: A: PDB Molecule: protein rv1364c/mt1410;PDBTitle: the crystal structure of the rsbu and rsbw domains of rv1364c from2 mycobacterium tuberculosis
23 c3es2A_
not modelled
98.7
12
PDB header: signaling proteinChain: A: PDB Molecule: probable two-component response regulator;PDBTitle: structure of the c-terminal phosphatase domain of p.2 aeruginonsa rssb
24 c3eq2A_
not modelled
28.8
11
PDB header: signaling proteinChain: A: PDB Molecule: probable two-component response regulator;PDBTitle: structure of hexagonal crystal form of pseudomonas2 aeruginosa rssb
25 c1rfoC_
not modelled
10.1
25
PDB header: viral proteinChain: C: PDB Molecule: whisker antigen control protein;PDBTitle: trimeric foldon of the t4 phagehead fibritin
26 c3iddA_
not modelled
6.7
20
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: cofactor-independent phosphoglycerate mutase from2 thermoplasma acidophilum dsm 1728
27 c3m8yC_
not modelled
5.7
25
PDB header: isomeraseChain: C: PDB Molecule: phosphopentomutase;PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
28 c3q4fG_
not modelled
5.6
14
PDB header: dna binding protein/protein bindingChain: G: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of xrcc4/xlf-cernunnos complex
29 c3mudA_
not modelled
5.4
14
PDB header: contractile proteinChain: A: PDB Molecule: dna repair protein xrcc4, tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
30 c2zktB_
not modelled
5.4
17
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
31 c3sr2A_
not modelled
5.4
14
PDB header: dna binding protein/protein bindingChain: A: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of human xlf-xrcc4 complex
32 d1ik9a1
not modelled
5.3
14
Fold: XRCC4, N-terminal domainSuperfamily: XRCC4, N-terminal domainFamily: XRCC4, N-terminal domain33 c1avyA_
not modelled
5.3
25
PDB header: coiled coilChain: A: PDB Molecule: fibritin;PDBTitle: fibritin deletion mutant m (bacteriophage t4)