1 c2v43A_
100.0
100
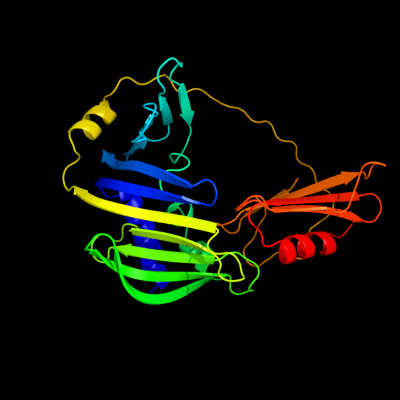
PDB header: regulatorChain: A: PDB Molecule: sigma-e factor regulatory protein rseb;PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
2 c3buuB_
99.9
11
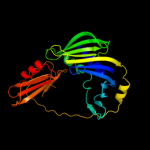
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized lola superfamily protein ne2245;PDBTitle: crystal structure of lola superfamily protein ne2245 from2 nitrosomonas europaea
3 c3bk5A_
99.9
13
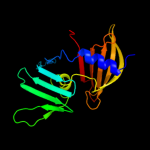
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative outer membrane lipoprotein-sorting protein;PDBTitle: crystal structure of putative outer membrane lipoprotein-sorting2 protein domain from vibrio parahaemolyticus
4 c2w7qB_
99.7
10
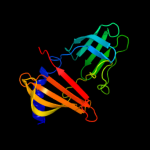
PDB header: protein transportChain: B: PDB Molecule: outer-membrane lipoprotein carrier protein;PDBTitle: structure of pseudomonas aeruginosa lola
5 d1iwla_
99.6
10
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factorsSuperfamily: Prokaryotic lipoproteins and lipoprotein localization factorsFamily: Outer-membrane lipoproteins carrier protein LolA6 c2yzyA_
98.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha1012;PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
7 c3mhaB_
98.1
14
PDB header: lipid binding proteinChain: B: PDB Molecule: lipoprotein lprg;PDBTitle: crystal structure of lprg from mycobacterium tuberculosis bound to pim
8 d2byoa1
97.1
16
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factorsSuperfamily: Prokaryotic lipoproteins and lipoprotein localization factorsFamily: LppX-like9 c3fzxA_
90.0
13
PDB header: lipid binding proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of a putative exported protein with ymcc-like fold2 (bf2203) from bacteroides fragilis nctc 9343 at 2.22 a resolution
10 c3mx7A_
77.1
18
PDB header: apoptosisChain: A: PDB Molecule: fas apoptotic inhibitory molecule 1;PDBTitle: crystal structure analysis of human faim-ntd
11 d2jnaa1
34.4
21
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like12 c3h4rA_
24.7
36
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease 8;PDBTitle: crystal structure of e. coli rece exonuclease
13 d1ei5a1
18.9
31
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains14 d1khia1
18.6
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like15 d1f15a_
18.4
15
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP16 c3m7aA_
17.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of saro_0823 (yp_496102.1) a protein of2 unknown function from novosphingobium aromaticivorans dsm3 12444 at 1.22 a resolution
17 c2vy8A_
12.7
15
PDB header: transcriptionChain: A: PDB Molecule: polymerase basic protein 2;PDBTitle: the 627-domain from influenza a virus polymerase pb22 subunit with glu-627
18 c2jcmA_
12.4
30
PDB header: hydrolaseChain: A: PDB Molecule: cytosolic purine 5'-nucleotidase;PDBTitle: crystal structure of human cytosolic 5'-nucleotidase ii in2 complex with beryllium trifluoride
19 d1fe0a_
12.0
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain20 d1u5da1
11.3
13
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)21 d1bcca1
not modelled
11.3
14
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like22 d2bdea1
not modelled
10.8
7
Fold: HAD-likeSuperfamily: HAD-likeFamily: 5' nucleotidase-like23 d3cx5a1
not modelled
10.7
18
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like24 c3rd4A_
not modelled
10.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of propen_03304 from proteus penneri atcc 351982 northeast structural genomics consortium target id pvr55
25 d1ufza_
not modelled
9.2
21
Fold: RuvA C-terminal domain-likeSuperfamily: HBS1-like domainFamily: HBS1-like domain26 c3pjyB_
not modelled
8.9
20
PDB header: transcription regulatorChain: B: PDB Molecule: hypothetical signal peptide protein;PDBTitle: crystal structure of a putative transcription regulator (r01717) from2 sinorhizobium meliloti 1021 at 1.55 a resolution
27 d1ci3m1
not modelled
8.3
42
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain28 c1jmtB_
not modelled
8.3
27
PDB header: rna binding proteinChain: B: PDB Molecule: splicing factor u2af 65 kda subunit;PDBTitle: x-ray structure of a core u2af65/u2af35 heterodimer
29 c2crlA_
not modelled
8.2
21
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
30 d1e2wa1
not modelled
8.2
42
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain31 c2opdA_
not modelled
8.0
20
PDB header: cell adhesionChain: A: PDB Molecule: pilx;PDBTitle: structure of the neisseria meningitidis minor type iv pilin,2 pilx
32 d1tu2b1
not modelled
8.0
42
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain33 d1hcza1
not modelled
7.9
42
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain34 d1vf5c1
not modelled
7.8
42
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain35 d1q2la1
not modelled
7.2
18
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like36 c2jxmB_
not modelled
7.1
42
PDB header: electron transportChain: B: PDB Molecule: cytochrome f;PDBTitle: ensemble of twenty structures of the prochlorothrix2 hollandica plastocyanin- cytochrome f complex
37 c3nqyA_
not modelled
7.0
9
PDB header: hydrolaseChain: A: PDB Molecule: secreted metalloprotease mcp02;PDBTitle: crystal structure of the autoprocessed complex of vibriolysin mcp-022 with a single point mutation e346a
38 c1tu2B_
not modelled
7.0
42
PDB header: electron transportChain: B: PDB Molecule: apocytochrome f;PDBTitle: the complex of nostoc cytochrome f and plastocyanin determin with2 paramagnetic nmr. based on the structures of cytochrome f and3 plastocyanin, 10 structures
39 c1ctmA_
not modelled
6.9
42
PDB header: electron transport(cytochrome)Chain: A: PDB Molecule: cytochrome f;PDBTitle: crystal structure of chloroplast cytochrome f reveals a2 novel cytochrome fold and unexpected heme ligation
40 c1e2vB_
not modelled
6.8
42
PDB header: electron transport proteinsChain: B: PDB Molecule: cytochrome f;PDBTitle: n153q mutant of cytochrome f from chlamydomonas reinhardtii
41 d1tafb_
not modelled
6.3
35
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs42 d1hr6b1
not modelled
5.9
14
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like43 c1q90A_
not modelled
5.7
42
PDB header: photosynthesisChain: A: PDB Molecule: apocytochrome f;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii