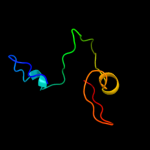
| 1 | d1zupa1 |
|
 |
51.0 |
38 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:TM1739-like |
| 2 | c2jlnA_ |
|
 |
38.1 |
28 |
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
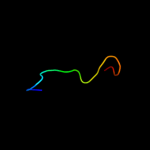
| 3 | c3mcuF_ |
|
 |
14.6 |
26 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dipicolinate synthase, b chain;
PDBTitle: crystal structure of the dipicolinate synthase chain b from2 bacillus cereus. northeast structural genomics consortium3 target bcr215.
|
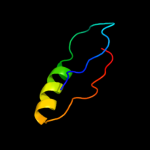
| 4 | c1zc1A_ |
|
 |
11.9 |
30 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
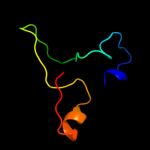
| 5 | c2yujA_ |
|
 |
10.9 |
36 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
|
| 6 | c3pvcA_ |
|
 |
8.9 |
26 |
PDB header:oxidoreductase, transferase
Chain: A: PDB Molecule:trna 5-methylaminomethyl-2-thiouridine biosynthesis
PDBTitle: crystal structure of apo mnmc from yersinia pestis
|
| 7 | c3lqkA_ |
|
 |
8.9 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
| 8 | c2l14B_ |
|
 |
6.7 |
41 |
PDB header:protein binding
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: structure of cbp nuclear coactivator binding domain in complex with2 p53 tad
|
| 9 | d1q3ja_ |
|
 |
6.5 |
50 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Gurmarin-like
Family:Antifungal peptide |
| 10 | d1c99a_ |
|
 |
5.3 |
86 |
Fold:Transmembrane helix hairpin
Superfamily:F1F0 ATP synthase subunit C
Family:F1F0 ATP synthase subunit C |

