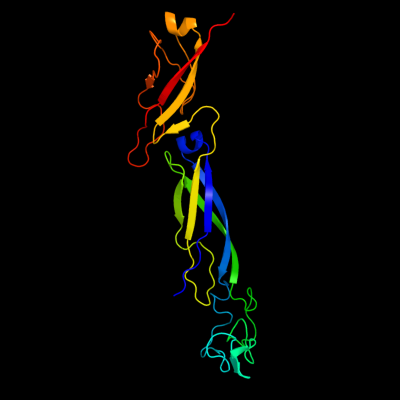
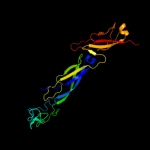
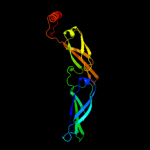
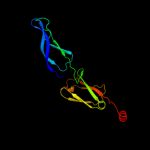
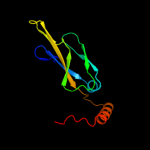
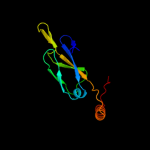
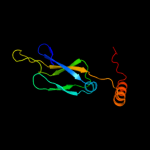
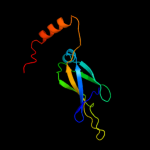
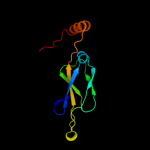
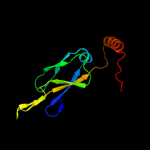
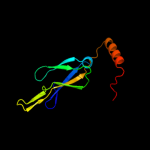
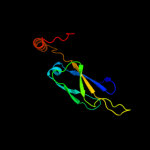
1 c1nltA_
100.0
28
PDB header: protein transportChain: A: PDB Molecule: mitochondrial protein import protein mas5;PDBTitle: the crystal structure of hsp40 ydj1
2 c3lz8A_
100.0
36
PDB header: chaperoneChain: A: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone dnaj from klebsiella pneumoniae2 subsp. pneumoniae mgh 78578 at 2.9 a resolution.
3 c2q2gA_
100.0
26
PDB header: chaperoneChain: A: PDB Molecule: heat shock 40 kda protein, putative (fragment);PDBTitle: crystal structure of dimerization domain of hsp40 from2 cryptosporidium parvum, cgd2_1800
4 c2qldA_
100.0
26
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 1;PDBTitle: human hsp40 hdj1
5 c2b26A_
100.0
25
PDB header: chaperone/protein transportChain: A: PDB Molecule: sis1 protein;PDBTitle: the crystal structure of the protein complex of yeast hsp402 sis1 and hsp70 ssa1
6 c3apqB_
99.9
49
PDB header: oxidoreductaseChain: B: PDB Molecule: dnaj homolog subfamily c member 10;PDBTitle: crystal structure of j-trx1 fragment of erdj5
7 c3i38D_
99.9
32
PDB header: chaperoneChain: D: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
8 c3i38K_
99.9
32
PDB header: chaperoneChain: K: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
9 c3i38A_
99.9
32
PDB header: chaperoneChain: A: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
10 c3i38G_
99.9
32
PDB header: chaperoneChain: G: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
11 c3i38F_
99.9
33
PDB header: chaperoneChain: F: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
12 c3i38J_
99.9
33
PDB header: chaperoneChain: J: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
13 c3i38B_
99.9
33
PDB header: chaperoneChain: B: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
14 c3i38H_
99.9
32
PDB header: chaperoneChain: H: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
15 c3i38C_
99.9
33
PDB header: chaperoneChain: C: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
16 c3i38E_
99.9
33
PDB header: chaperoneChain: E: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
17 c2l6lA_
99.9
24
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 24;PDBTitle: solution structure of human j-protein co-chaperone, dph4
18 c3i38L_
99.9
33
PDB header: chaperoneChain: L: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
19 c3i38I_
99.9
32
PDB header: chaperoneChain: I: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
20 c2ctqA_
99.9
28
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 12;PDBTitle: solution structure of j-domain from human dnaj subfamily c2 menber 12
21 c2yuaA_
not modelled
99.9
38
PDB header: chaperoneChain: A: PDB Molecule: williams-beuren syndrome chromosome region 18PDBTitle: solution structure of the dnaj domain from human williams-2 beuren syndrome chromosome region 18 protein
22 d1c3ga2
not modelled
99.9
27
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain23 d1hdja_
99.9
50
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain24 c2ctwA_
99.9
45
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 5;PDBTitle: solution structure of j-domain from mouse dnaj subfamily c2 menber 5
25 c1fpoA_
not modelled
99.9
20
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein hscb;PDBTitle: hsc20 (hscb), a j-type co-chaperone from e. coli
26 d1xbla_
99.9
100
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain27 c3hhoA_
not modelled
99.9
22
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb homolog;PDBTitle: chaperone hscb from vibrio cholerae
28 c2dmxA_
not modelled
99.9
48
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 8;PDBTitle: solution structure of the j domain of dnaj homolog2 subfamily b member 8
29 c3bvoA_
not modelled
99.9
17
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb, mitochondrial precursor;PDBTitle: crystal structure of human co-chaperone protein hscb
30 c2ctrA_
not modelled
99.9
54
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 9;PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 9
31 c1bq0A_
99.8
100
PDB header: chaperoneChain: A: PDB Molecule: dnaj;PDBTitle: j-domain (residues 1-77) of the escherichia coli n-terminal2 fragment (residues 1-104) of the molecular chaperone dnaj,3 nmr, 20 structures
32 c2o37A_
not modelled
99.8
50
PDB header: chaperoneChain: A: PDB Molecule: protein sis1;PDBTitle: j-domain of sis1 protein, hsp40 co-chaperone from2 saccharomyces cerevisiae.
33 c1xaoA_
not modelled
99.8
24
PDB header: chaperoneChain: A: PDB Molecule: mitochondrial protein import protein mas5;PDBTitle: hsp40-ydj1 dimerization domain
34 c2ctpA_
not modelled
99.8
49
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 12;PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 12
35 c2kqxA_
not modelled
99.8
53
PDB header: chaperone binding proteinChain: A: PDB Molecule: curved dna-binding protein;PDBTitle: nmr structure of the j-domain (residues 2-72) in the2 escherichia coli cbpa
36 c2cugA_
not modelled
99.8
48
PDB header: chaperoneChain: A: PDB Molecule: mkiaa0962 protein;PDBTitle: solution structure of the j domain of the pseudo dnaj2 protein, mouse hypothetical mkiaa0962
37 c2dn9A_
not modelled
99.8
52
PDB header: apoptosis, chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
38 c2qsaA_
not modelled
99.8
36
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog dnj-2;PDBTitle: crystal structure of j-domain of dnaj homolog dnj-2 precursor from2 c.elegans.
39 d1gh6a_
not modelled
99.8
26
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain40 d1iura_
not modelled
99.8
25
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain41 c2ochA_
not modelled
99.8
48
PDB header: chaperoneChain: A: PDB Molecule: hypothetical protein dnj-12;PDBTitle: j-domain of dnj-12 from caenorhabditis elegans
42 d1wjza_
not modelled
99.8
30
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain43 c2ys8A_
not modelled
99.8
34
PDB header: protein bindingChain: A: PDB Molecule: rab-related gtp-binding protein rabj;PDBTitle: solution structure of the dnaj-like domain from human ras-2 associated protein rap1
44 d1nlta2
not modelled
99.8
28
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain45 c2pf4E_
not modelled
99.7
27
PDB header: hydrolase regulator/viral proteinChain: E: PDB Molecule: small t antigen;PDBTitle: crystal structure of the full-length simian virus 40 small t antigen2 complexed with the protein phosphatase 2a aalpha subunit
46 d1fpoa1
not modelled
99.7
25
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain47 d1fafa_
not modelled
99.6
17
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain48 d1n4ca_
not modelled
99.6
21
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain49 c2guzO_
not modelled
99.6
17
PDB header: chaperone, protein transportChain: O: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
50 d1nz6a_
not modelled
99.5
21
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain51 c2y4tA_
not modelled
99.4
52
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 3;PDBTitle: crystal structure of the human co-chaperone p58(ipk)
52 c2cttA_
not modelled
99.4
39
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of zinc finger domain from human dnaj2 subfamily a menber 3
53 d1c3ga1
not modelled
99.4
20
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain54 c3apoA_
not modelled
99.3
67
PDB header: oxidoreductaseChain: A: PDB Molecule: dnaj homolog subfamily c member 10;PDBTitle: crystal structure of full-length erdj5
55 c3ag7A_
not modelled
99.3
25
PDB header: plant proteinChain: A: PDB Molecule: putative uncharacterized protein f9e10.5;PDBTitle: an auxilin-like j-domain containing protein, jac1 j-domain
56 c2guzD_
not modelled
99.2
13
PDB header: chaperone, protein transportChain: D: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
57 d1nlta1
not modelled
99.2
13
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain58 d1exka_
not modelled
98.9
100
Fold: DnaJ/Hsp40 cysteine-rich domainSuperfamily: DnaJ/Hsp40 cysteine-rich domainFamily: DnaJ/Hsp40 cysteine-rich domain59 c3uo2A_
not modelled
98.9
25
PDB header: chaperoneChain: A: PDB Molecule: j-type co-chaperone jac1, mitochondrial;PDBTitle: jac1 co-chaperone from saccharomyces cerevisiae
60 d1nlta3
not modelled
98.2
32
Fold: DnaJ/Hsp40 cysteine-rich domainSuperfamily: DnaJ/Hsp40 cysteine-rich domainFamily: DnaJ/Hsp40 cysteine-rich domain61 c3ld0Q_
not modelled
85.7
43
PDB header: gene regulationChain: Q: PDB Molecule: inhibitor of trap, regulated by t-box (trp) sequence rtpa;PDBTitle: crystal structure of b.licheniformis anti-trap protein, an antagonist2 of trap-rna interactions
62 c2k3vA_
not modelled
80.6
25
PDB header: electron transportChain: A: PDB Molecule: tetraheme cytochrome c-type;PDBTitle: solution structure of a tetrahaem cytochrome from2 shewanella frigidimarina
63 c2bx9J_
not modelled
67.1
32
PDB header: transcription regulationChain: J: PDB Molecule: tryptophan rna-binding attenuator protein-inhibitoryPDBTitle: crystal structure of b.subtilis anti-trap protein, an2 antagonist of trap-rna interactions
64 d2q37a1
not modelled
56.5
20
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like65 c2q37A_
not modelled
56.5
20
PDB header: plant protein, lyaseChain: A: PDB Molecule: ohcu decarboxylase;PDBTitle: crystal structure of ohcu decarboxylase in the presence of2 (s)-allantoin
66 d1ug2a_
not modelled
52.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain67 d1akha_
not modelled
47.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain68 c3epvB_
not modelled
46.8
21
PDB header: metal binding proteinChain: B: PDB Molecule: nickel and cobalt resistance protein cnrr;PDBTitle: x-ray structure of the metal-sensor cnrx in both the apo- and copper-2 bound forms
69 c3dkqB_
not modelled
46.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: pkhd-type hydroxylase sbal_3634;PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
70 c1wz6A_
not modelled
45.2
20
PDB header: transcriptionChain: A: PDB Molecule: hmg-box transcription factor bbx;PDBTitle: solution structure of the hmg_box domain of murine bobby2 sox homolog
71 d2o8ia1
not modelled
43.6
23
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like72 d3saka_
not modelled
43.3
42
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain73 c2cs1A_
not modelled
42.3
13
PDB header: dna binding proteinChain: A: PDB Molecule: pms1 protein homolog 1;PDBTitle: solution structure of the hmg domain of human dna mismatch2 repair protein
74 c2da7A_
not modelled
39.7
17
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger homeobox protein 1b;PDBTitle: solution structure of the homeobox domain of zinc finger2 homeobox protein 1b (smad interacting protein 1)
75 c2co9A_
not modelled
39.6
13
PDB header: transcriptionChain: A: PDB Molecule: thymus high mobility group box protein tox;PDBTitle: solution structure of the hmg_box domain of thymus high2 mobility group box protein tox from mouse
76 c2crjA_
not modelled
39.6
15
PDB header: gene regulationChain: A: PDB Molecule: swi/snf-related matrix-associated actin-PDBTitle: solution structure of the hmg domain of mouse hmg domain2 protein hmgx2
77 c3m1gC_
not modelled
37.8
25
PDB header: transferaseChain: C: PDB Molecule: putative glutathione s-transferase;PDBTitle: the structure of a putative glutathione s-transferase from2 corynebacterium glutamicum
78 d2ecca1
not modelled
37.6
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain79 c1mszA_
not modelled
37.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein smubp-2;PDBTitle: solution structure of the r3h domain from human smubp-2
80 d1msza_
not modelled
37.0
25
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain81 d2o70a1
not modelled
35.3
23
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like82 d1o4xa1
not modelled
35.2
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain83 c3nv4A_
not modelled
33.8
9
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin 9 short isoform variant;PDBTitle: crystal structure of human galectin-9 c-terminal crd in complex with2 sialyllactose
84 c2e6oA_
not modelled
32.8
11
PDB header: transcription, cell cycleChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: solution structure of the hmg box domain from human hmg-box2 transcription factor 1
85 d3c7bb2
not modelled
32.2
21
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like86 c3pihA_
not modelled
31.3
28
PDB header: hydrolase/dnaChain: A: PDB Molecule: uvrabc system protein a;PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
87 d1wgfa_
not modelled
31.1
11
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box88 c2jucA_
not modelled
29.4
14
PDB header: unknown functionChain: A: PDB Molecule: pre-mrna-splicing factor urn1;PDBTitle: urn1 ff domain yeast
89 c3u2bC_
not modelled
29.3
20
PDB header: transcription/dnaChain: C: PDB Molecule: transcription factor sox-4;PDBTitle: structure of the sox4 hmg domain bound to dna
90 c2j11D_
not modelled
29.2
42
PDB header: transcriptionChain: D: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant y327s t329g q331g
91 d1aiea_
not modelled
28.8
42
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain92 c2ww9K_
not modelled
28.7
47
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l25;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
93 c3c4rC_
not modelled
28.5
28
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
94 c2j10B_
not modelled
28.1
42
PDB header: transcriptionChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
95 c2j10A_
not modelled
28.1
42
PDB header: transcriptionChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
96 c2j10D_
not modelled
28.1
42
PDB header: transcriptionChain: D: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
97 d1vqos1
not modelled
27.8
24
Fold: Ribosomal proteins S24e, L23 and L15eSuperfamily: Ribosomal proteins S24e, L23 and L15eFamily: L23p98 d1le8a_
not modelled
27.6
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain99 d1mh3a1
not modelled
27.5
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain100 c3narA_
not modelled
27.3
15
PDB header: transcriptionChain: A: PDB Molecule: zinc fingers and homeoboxes protein 1;PDBTitle: crystal structure of zhx1 hd4 (zinc-fingers and homeoboxes protein 1,2 homeodomain 4)
101 d2cqna1
not modelled
27.1
16
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain102 c2yroA_
not modelled
26.9
9
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: solution structure of the c-terminal gal-bind lectin2 protein from human galectin-8
103 c2bl0B_
not modelled
26.9
21
PDB header: muscle proteinChain: B: PDB Molecule: myosin regulatory light chain;PDBTitle: physarum polycephalum myosin ii regulatory domain
104 d1uzca_
not modelled
26.6
23
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain105 c2da2A_
not modelled
26.6
15
PDB header: transcriptionChain: A: PDB Molecule: alpha-fetoprotein enhancer binding protein;PDBTitle: solution structure of the second homeobox domain of at-2 binding transcription factor 1 (atbf1)
106 d2j0td1
not modelled
26.4
16
Fold: OB-foldSuperfamily: TIMP-likeFamily: Tissue inhibitor of metalloproteinases, TIMP107 c3fn2A_
not modelled
26.4
25
PDB header: transferaseChain: A: PDB Molecule: putative sensor histidine kinase domain;PDBTitle: crystal structure of a putative sensor histidine kinase domain from2 clostridium symbiosum atcc 14940
108 c1x50A_
not modelled
25.9
14
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-4;PDBTitle: solution structure of the c-terminal gal-bind lectin domain2 from human galectin-4
109 d1e3oc1
not modelled
25.6
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain110 c2go54_
not modelled
24.8
29
PDB header: translation/rnaChain: 4: PDB Molecule: ribosomal protein l23;PDBTitle: structure of signal recognition particle receptor (sr) in2 complex with signal recognition particle (srp) and3 ribosome nascent chain complex
111 d2fiya1
not modelled
24.6
18
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like112 c2da3A_
not modelled
24.5
15
PDB header: transcriptionChain: A: PDB Molecule: alpha-fetoprotein enhancer binding protein;PDBTitle: solution structure of the third homeobox domain of at-2 binding transcription factor 1 (atbf1)
113 c2dycA_
not modelled
24.1
19
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-4;PDBTitle: crystal structure of the n-terminal domain of mouse galectin-4
114 d1h8ba_
not modelled
24.1
17
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins115 d1gefa_
not modelled
23.9
24
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like116 d1cf7a_
not modelled
23.9
44
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp117 d1gyxa_
not modelled
22.7
24
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like118 c2zkrs_
not modelled
22.6
35
PDB header: ribosomal protein/rnaChain: S: PDB Molecule: rna expansion segment es39 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
119 c3c7bE_
not modelled
22.5
21
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
120 d1hcza1
not modelled
22.3
22
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain