| 1 |
|
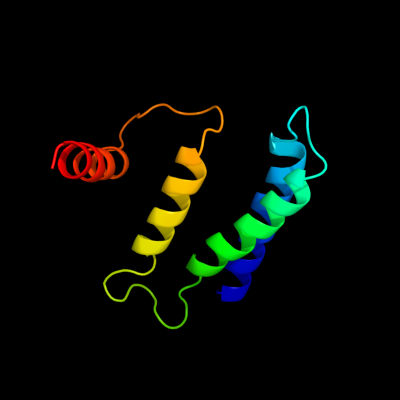
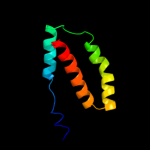
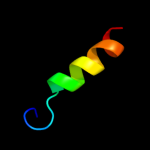
PDB 1s7b chain A
Region: 9 - 101
Aligned: 93
Modelled: 93
Confidence: 95.6%
Identity: 25%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
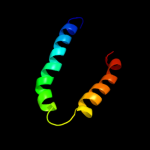
PDB 1pv7 chain A
Region: 51 - 103
Aligned: 53
Modelled: 53
Confidence: 20.1%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
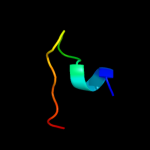
PDB 1pw4 chain A
Region: 51 - 103
Aligned: 53
Modelled: 53
Confidence: 16.1%
Identity: 15%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 4 |
|
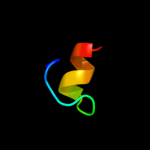
PDB 2xut chain C
Region: 51 - 103
Aligned: 53
Modelled: 53
Confidence: 12.6%
Identity: 15%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 5 |
|
PDB 1w8x chain P
Region: 21 - 28
Aligned: 8
Modelled: 8
Confidence: 10.3%
Identity: 50%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 6 |
|
PDB 3o7p chain A
Region: 30 - 103
Aligned: 74
Modelled: 74
Confidence: 10.3%
Identity: 18%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 7 |
|
PDB 2ipq chain X domain 1
Region: 19 - 29
Aligned: 11
Modelled: 11
Confidence: 8.2%
Identity: 36%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: STY4665 C-terminal domain-like
Phyre2
| 8 |
|
PDB 3msq chain C
Region: 48 - 63
Aligned: 16
Modelled: 16
Confidence: 7.3%
Identity: 38%
PDB header:biosynthetic protein
Chain: C: PDB Molecule:putative ubiquinone biosynthesis protein;
PDBTitle: crystal structure of a putative ubiquinone biosynthesis protein2 (npun02000094) from nostoc punctiforme pcc 73102 at 2.85 a resolution
Phyre2
| 9 |
|
PDB 1b1y chain A
Region: 78 - 90
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 38%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 10 |
|
PDB 6rlx chain B
Region: 76 - 99
Aligned: 24
Modelled: 24
Confidence: 6.5%
Identity: 33%
PDB header:hormone(muscle relaxant)
Chain: B: PDB Molecule:relaxin, b-chain;
PDBTitle: x-ray structure of human relaxin at 1.5 angstroms. comparison to2 insulin and implications for receptor binding determinants
Phyre2
| 11 |
|
PDB 2kbv chain A
Region: 62 - 81
Aligned: 20
Modelled: 20
Confidence: 6.1%
Identity: 30%
PDB header:membrane protein
Chain: A: PDB Molecule:sodium/hydrogen exchanger 1;
PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
Phyre2
| 12 |
|
PDB 2gfp chain A
Region: 51 - 103
Aligned: 53
Modelled: 53
Confidence: 6.1%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 13 |
|
PDB 3dve chain B
Region: 55 - 62
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 63%
PDB header:membrane protein
Chain: B: PDB Molecule:voltage-dependent n-type calcium channel subunit alpha-1b;
PDBTitle: crystal structure of ca2+/cam-cav2.2 iq domain complex
Phyre2
| 14 |
|
PDB 2xfy chain A
Region: 74 - 90
Aligned: 17
Modelled: 17
Confidence: 5.7%
Identity: 35%
PDB header:hydrolase
Chain: A: PDB Molecule:beta-amylase;
PDBTitle: crystal structure of barley beta-amylase complexed with2 alpha-cyclodextrin
Phyre2
| 15 |
|
PDB 1wdp chain A domain 1
Region: 74 - 90
Aligned: 17
Modelled: 17
Confidence: 5.7%
Identity: 24%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 16 |
|
PDB 3kb4 chain D
Region: 48 - 63
Aligned: 16
Modelled: 16
Confidence: 5.5%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:alr8543 protein;
PDBTitle: crystal structure of the alr8543 protein in complex with2 geranylgeranyl monophosphate and magnesium ion from nostoc sp. pcc3 7120, northeast structural genomics consortium target nsr141
Phyre2
| 17 |
|
PDB 1fa2 chain A
Region: 74 - 90
Aligned: 17
Modelled: 17
Confidence: 5.1%
Identity: 24%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 18 |
|
PDB 2fno chain A domain 1
Region: 68 - 85
Aligned: 18
Modelled: 18
Confidence: 5.1%
Identity: 28%
Fold: GST C-terminal domain-like
Superfamily: GST C-terminal domain-like
Family: Glutathione S-transferase (GST), C-terminal domain
Phyre2