| 1 |
|
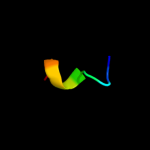
PDB 3h92 chain A
Region: 28 - 38
Aligned: 11
Modelled: 11
Confidence: 24.0%
Identity: 55%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized atp-binding protein mjecl15;
PDBTitle: the crystal structure of one domain of the protein with unknown2 function from methanocaldococcus jannaschii
Phyre2
| 2 |
|
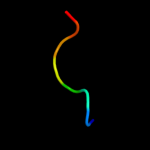
PDB 3pvd chain A
Region: 27 - 45
Aligned: 18
Modelled: 19
Confidence: 22.4%
Identity: 22%
PDB header:viral protein
Chain: A: PDB Molecule:capsid;
PDBTitle: crystal structure of p domain dimer of norovirus va207 complexed with2 3'-sialyl-lewis x tetrasaccharide
Phyre2
| 3 |
|
PDB 2zl5 chain A
Region: 27 - 45
Aligned: 18
Modelled: 19
Confidence: 20.4%
Identity: 39%
PDB header:viral protein
Chain: A: PDB Molecule:58 kd capsid protein;
PDBTitle: atomic resolution structural characterization of2 recognition of histo-blood group antigen by norwalk virus
Phyre2
| 4 |
|
PDB 3pm5 chain B
Region: 10 - 31
Aligned: 22
Modelled: 22
Confidence: 20.1%
Identity: 32%
PDB header:oxidoreductase
Chain: B: PDB Molecule:benzoyl-coa oxygenase component b;
PDBTitle: crystal structure of boxb in mixed valent state with bound benzoyl-coa
Phyre2
| 5 |
|
PDB 3lq6 chain A
Region: 27 - 45
Aligned: 18
Modelled: 19
Confidence: 17.8%
Identity: 39%
PDB header:viral protein
Chain: A: PDB Molecule:capsid protein;
PDBTitle: crystal structure of murine norovirus protruding (p) domain
Phyre2
| 6 |
|
PDB 2obt chain A
Region: 27 - 45
Aligned: 18
Modelled: 19
Confidence: 16.8%
Identity: 28%
PDB header:viral protein
Chain: A: PDB Molecule:capsid protein;
PDBTitle: crystal structures of p domain of norovirus va387 in2 complex with blood group trisaccharides type b
Phyre2
| 7 |
|
PDB 2vy2 chain A
Region: 57 - 66
Aligned: 10
Modelled: 10
Confidence: 16.8%
Identity: 40%
PDB header:transcription
Chain: A: PDB Molecule:protein leafy;
PDBTitle: structure of leafy transcription factor from arabidopsis2 thaliana in complex with dna from ag-i promoter
Phyre2
| 8 |
|
PDB 3g6i chain A
Region: 16 - 22
Aligned: 7
Modelled: 7
Confidence: 13.8%
Identity: 100%
PDB header:unknown function
Chain: A: PDB Molecule:putative outer membrane protein, part of carbohydrate
PDBTitle: crystal structure of an outer membrane protein, part of a putative2 carbohydrate binding complex (bt_1022) from bacteroides3 thetaiotaomicron vpi-5482 at 1.93 a resolution
Phyre2
| 9 |
|
PDB 1mqr chain A
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 13.7%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-d-glucuronidase;
PDBTitle: the crystal structure of alpha-d-glucuronidase (e386q) from bacillus2 stearothermophilus t-6
Phyre2
| 10 |
|
PDB 1l8n chain A domain 1
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 13.6%
Identity: 50%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: alpha-D-glucuronidase/Hyaluronidase catalytic domain
Phyre2
| 11 |
|
PDB 3bjo chain A
Region: 28 - 38
Aligned: 11
Modelled: 11
Confidence: 13.0%
Identity: 45%
PDB header:nucleotide binding protein
Chain: A: PDB Molecule:uncharacterized atp-binding protein mj1010;
PDBTitle: crystal structure of the c-terminal domain of a possible atp-binding2 protein from methanocaldococcus jannaschii dsm 2661
Phyre2
| 12 |
|
PDB 2hl2 chain A
Region: 3 - 27
Aligned: 25
Modelled: 25
Confidence: 12.8%
Identity: 40%
PDB header:ligase
Chain: A: PDB Molecule:threonyl-trna synthetase;
PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase from pyrococcus abyssi in complex with an3 analog of seryladenylate
Phyre2
| 13 |
|
PDB 2i5n chain H domain 2
Region: 19 - 37
Aligned: 19
Modelled: 19
Confidence: 9.0%
Identity: 32%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 14 |
|
PDB 1w36 chain C domain 3
Region: 28 - 71
Aligned: 44
Modelled: 44
Confidence: 8.9%
Identity: 23%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Exodeoxyribonuclease V beta chain (RecC), C-terminal domain
Phyre2
| 15 |
|
PDB 3cjr chain B domain 1
Region: 1 - 15
Aligned: 15
Modelled: 15
Confidence: 8.0%
Identity: 47%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Ribosomal protein L11, C-terminal domain
Family: Ribosomal protein L11, C-terminal domain
Phyre2
| 16 |
|
PDB 1h41 chain A domain 1
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 7.6%
Identity: 50%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: alpha-D-glucuronidase/Hyaluronidase catalytic domain
Phyre2
| 17 |
|
PDB 1gqk chain B
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 7.3%
Identity: 50%
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-d-glucuronidase;
PDBTitle: structure of pseudomonas cellulosa alpha-d-glucuronidase2 complexed with glucuronic acid
Phyre2
| 18 |
|
PDB 3q38 chain A
Region: 27 - 43
Aligned: 16
Modelled: 17
Confidence: 7.2%
Identity: 19%
PDB header:viral protein
Chain: A: PDB Molecule:capsid protein;
PDBTitle: crystal structure of p domain from norwalk virus strain vietnam 026 in2 complex with hbga type b (triglycan)
Phyre2
| 19 |
|
PDB 1zto chain A
Region: 10 - 17
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 50%
PDB header:potassium channel
Chain: A: PDB Molecule:potassium channel protein rck4;
PDBTitle: inactivation gate of potassium channel rck4, nmr, 82 structures
Phyre2
| 20 |
|
PDB 2yzj chain B
Region: 17 - 25
Aligned: 9
Modelled: 9
Confidence: 5.7%
Identity: 67%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:167aa long hypothetical dutpase;
PDBTitle: crystal structure of dctp deaminase from sulfolobus tokodaii
Phyre2
| 21 |
|
| 22 |
|