| 1 |
|
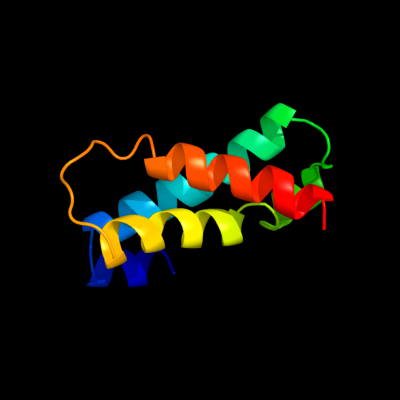
PDB 3k3g chain A
Region: 32 - 112
Aligned: 80
Modelled: 81
Confidence: 27.5%
Identity: 24%
PDB header:transport protein
Chain: A: PDB Molecule:urea transporter;
PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
Phyre2
| 2 |
|
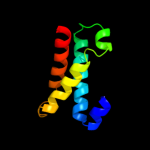
PDB 1iwg chain A domain 8
Region: 1 - 89
Aligned: 85
Modelled: 89
Confidence: 23.5%
Identity: 9%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 3 |
|
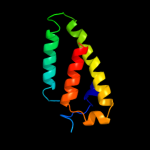
PDB 1oy8 chain A
Region: 14 - 89
Aligned: 72
Modelled: 76
Confidence: 16.5%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 4 |
|
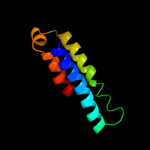
PDB 2knc chain A
Region: 93 - 108
Aligned: 16
Modelled: 16
Confidence: 13.2%
Identity: 25%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 5 |
|
PDB 2l0e chain A
Region: 279 - 292
Aligned: 14
Modelled: 14
Confidence: 11.4%
Identity: 29%
PDB header:membrane protein
Chain: A: PDB Molecule:sodium/hydrogen exchanger 1;
PDBTitle: structural and functional analysis of tm vi of the nhe1 isoform of the2 na+/h+ exchanger
Phyre2
| 6 |
|
PDB 3knu chain D
Region: 327 - 351
Aligned: 25
Modelled: 25
Confidence: 7.7%
Identity: 24%
PDB header:transferase
Chain: D: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna (guanine-n1)-methyltransferase from2 anaplasma phagocytophilum
Phyre2
| 7 |
|
PDB 1jb7 chain A domain 3
Region: 114 - 122
Aligned: 9
Modelled: 9
Confidence: 6.6%
Identity: 67%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Single strand DNA-binding domain, SSB
Phyre2
| 8 |
|
PDB 1ymg chain A
Region: 79 - 125
Aligned: 47
Modelled: 47
Confidence: 5.4%
Identity: 21%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 9 |
|
PDB 1ymg chain A domain 1
Region: 79 - 125
Aligned: 47
Modelled: 47
Confidence: 5.4%
Identity: 21%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 10 |
|
PDB 2axt chain J domain 1
Region: 93 - 113
Aligned: 21
Modelled: 21
Confidence: 5.3%
Identity: 29%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2