| 1 |
|
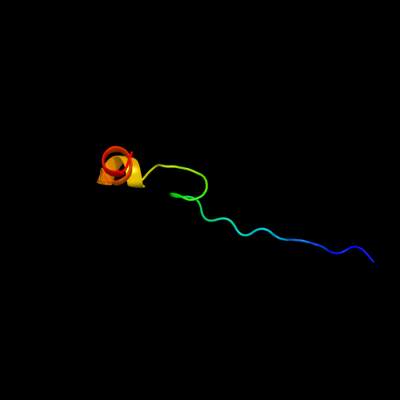
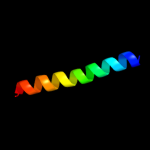
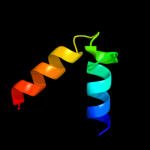
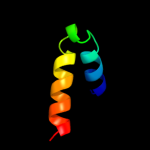
PDB 3geb chain C
Region: 18 - 47
Aligned: 30
Modelled: 30
Confidence: 52.2%
Identity: 30%
PDB header:hydrolase
Chain: C: PDB Molecule:eyes absent homolog 2;
PDBTitle: crystal structure of edeya2
Phyre2
| 2 |
|
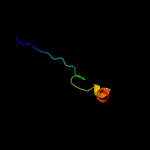
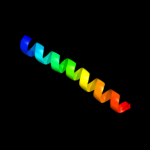
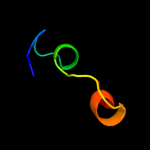
PDB 1ttz chain A
Region: 10 - 31
Aligned: 22
Modelled: 22
Confidence: 30.0%
Identity: 23%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Thioltransferase
Phyre2
| 3 |
|
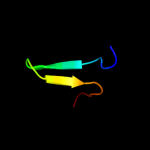
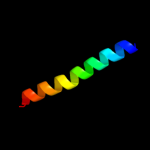
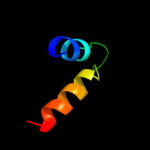
PDB 3d35 chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 14.9%
Identity: 25%
PDB header:transferase
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: crystal structure of rtt109-ac-coa complex
Phyre2
| 4 |
|
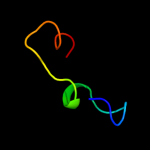
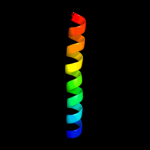
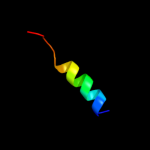
PDB 3a0b chain X
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 14.9%
Identity: 21%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 5 |
|
PDB 3a0h chain X
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 14.9%
Identity: 21%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 6 |
|
PDB 3a0b chain X
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 14.9%
Identity: 21%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 7 |
|
PDB 3a0h chain X
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 14.9%
Identity: 21%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 8 |
|
PDB 2zfn chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 13.4%
Identity: 25%
PDB header:transferase
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: self-acetylation mediated histone h3 lysine 56 acetylation by rtt109
Phyre2
| 9 |
|
PDB 1s5l chain X
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 13.1%
Identity: 21%
PDB header:photosynthesis
Chain: X: PDB Molecule:photosystem ii psbx protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 10 |
|
PDB 2y3a chain A
Region: 54 - 85
Aligned: 29
Modelled: 32
Confidence: 11.9%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: crystal structure of p110beta in complex with icsh2 of p85beta and2 the drug gdc-0941
Phyre2
| 11 |
|
PDB 3cz7 chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 11.6%
Identity: 25%
PDB header:replication
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: molecular basis for the autoregulation of the protein acetyl2 transferase rtt109
Phyre2
| 12 |
|
PDB 2wxo chain A
Region: 56 - 85
Aligned: 27
Modelled: 30
Confidence: 10.1%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: the crystal structure of the murine class ia pi 3-kinase2 p110delta in complex with as5.
Phyre2
| 13 |
|
PDB 2rlf chain A
Region: 73 - 90
Aligned: 18
Modelled: 18
Confidence: 8.8%
Identity: 22%
PDB header:proton transport
Chain: A: PDB Molecule:matrix protein 2;
PDBTitle: proton channel m2 from influenza a in complex with2 inhibitor rimantadine
Phyre2
| 14 |
|
PDB 2ysc chain A domain 1
Region: 78 - 83
Aligned: 6
Modelled: 6
Confidence: 8.5%
Identity: 50%
Fold: WW domain-like
Superfamily: WW domain
Family: WW domain
Phyre2
| 15 |
|
PDB 2rd0 chain A
Region: 56 - 85
Aligned: 27
Modelled: 30
Confidence: 8.2%
Identity: 22%
PDB header:transferase/oncoprotein
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: structure of a human p110alpha/p85alpha complex
Phyre2
| 16 |
|
PDB 2qvw chain A
Region: 3 - 23
Aligned: 21
Modelled: 21
Confidence: 7.6%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:glp_546_48378_50642;
PDBTitle: structure of giardia dicer refined against twinned data
Phyre2
| 17 |
|
PDB 1e8z chain A
Region: 56 - 87
Aligned: 29
Modelled: 32
Confidence: 6.5%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol 3-kinase catalytic subunit;
PDBTitle: structure determinants of phosphoinositide 3-kinase2 inhibition by wortmannin, ly294002, quercetin, myricetin3 and staurosporine
Phyre2
| 18 |
|
PDB 2ho2 chain A domain 1
Region: 78 - 83
Aligned: 6
Modelled: 6
Confidence: 6.1%
Identity: 67%
Fold: WW domain-like
Superfamily: WW domain
Family: WW domain
Phyre2
| 19 |
|
PDB 1fw5 chain A
Region: 79 - 83
Aligned: 5
Modelled: 5
Confidence: 6.0%
Identity: 40%
PDB header:viral protein
Chain: A: PDB Molecule:nonstructural protein nsp1;
PDBTitle: solution structure of membrane binding peptide of semliki2 forest virus mrna capping enzyme nsp1
Phyre2
| 20 |
|
PDB 1e7u chain A domain 1
Region: 54 - 87
Aligned: 31
Modelled: 34
Confidence: 5.1%
Identity: 13%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: Phoshoinositide 3-kinase (PI3K) helical domain
Phyre2
| 21 |
|