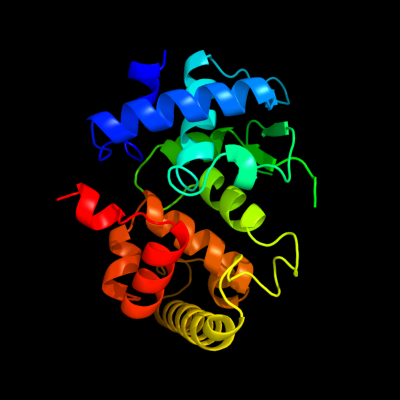
| 1 | c3dl1A_
|
|
 |
100.0 |
80 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent hydrolase;
PDBTitle: crystal structure of a putative metal-dependent hydrolase2 (yp_001336084.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 2.20 a resolution
|
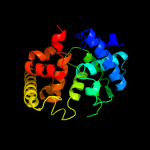
| 2 | c2l0rA_
|
|
 |
99.8 |
18 |
PDB header:hydrolase,toxin
Chain: A: PDB Molecule:lethal factor;
PDBTitle: conformational dynamics of the anthrax lethal factor catalytic center
|
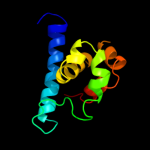
| 3 | d1j7na2
|
|
 |
99.8 |
16 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Anthrax toxin lethal factor, N- and C-terminal domains |
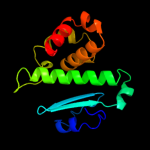
| 4 | c1yqyA_
|
|
 |
97.2 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:lethal factor;
PDBTitle: structure of b. anthrax lethal factor in complex with a2 hydroxamate inhibitor
|
| 5 | c1pwqA_
|
|
 |
97.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:lethal factor;
PDBTitle: crystal structure of anthrax lethal factor complexed with2 thioacetyl-tyr-pro-met-amide, a metal-chelating peptidyl3 small molecule inhibitor
|
| 6 | d2ejqa1
|
|
 |
84.6 |
13 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TTHA0227-like |
| 7 | c2xpyA_
|
|
 |
64.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:leukotriene a-4 hydrolase;
PDBTitle: structure of native leukotriene a4 hydrolase from saccharomyces2 cerevisiae
|
| 8 | c3lq0A_
|
|
 |
59.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:proastacin;
PDBTitle: zymogen structure of crayfish astacin metallopeptidase
|
| 9 | d1hv5a_
|
|
 |
57.3 |
29 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 10 | d1qiba_
|
|
 |
55.8 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 11 | d1i76a_
|
|
 |
53.6 |
42 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 12 | d1k7ia2
|
|
 |
53.2 |
23 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Serralysin-like metalloprotease, catalytic (N-terminal) domain |
| 13 | d1hfca_
|
|
 |
53.1 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 14 | d1kapp2
|
|
 |
52.4 |
31 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Serralysin-like metalloprotease, catalytic (N-terminal) domain |
| 15 | d1q3aa_
|
|
 |
52.2 |
42 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 16 | d1asta_
|
|
 |
51.5 |
50 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Astacin |
| 17 | d1mmqa_
|
|
 |
51.2 |
29 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 18 | d1cxva_
|
|
 |
50.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 19 | c3edhA_
|
|
 |
50.3 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:bone morphogenetic protein 1;
PDBTitle: crystal structure of bone morphogenetic protein 1 protease2 domain in complex with partially bound dmso
|
| 20 | c3ciaA_
|
|
 |
48.9 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:cold-active aminopeptidase;
PDBTitle: crystal structure of cold-aminopeptidase from colwellia2 psychrerythraea
|
| 21 | d1cgla_ |
|
not modelled |
48.5 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 22 | d1y93a1 |
|
not modelled |
48.0 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 23 | d2ixoa1 |
|
not modelled |
48.0 |
15 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 24 | d1g9ka2 |
|
not modelled |
47.8 |
22 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Serralysin-like metalloprotease, catalytic (N-terminal) domain |
| 25 | c3lqbA_ |
|
not modelled |
47.4 |
40 |
PDB header:hydrolase
Chain: A: PDB Molecule:loc792177 protein;
PDBTitle: crystal structure of the hatching enzyme zhe1 from the zebrafish danio2 rerio
|
| 26 | d1hy7a_ |
|
not modelled |
47.3 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 27 | d1bqqm_ |
|
not modelled |
46.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 28 | d1fbla2 |
|
not modelled |
45.6 |
28 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 29 | c2xs4A_ |
|
not modelled |
45.6 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:karilysin protease;
PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
|
| 30 | d1eaka2 |
|
not modelled |
45.5 |
15 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 31 | c2jsdA_ |
|
not modelled |
44.9 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:matrix metalloproteinase-20;
PDBTitle: solution structure of mmp20 complexed with nngh
|
| 32 | d1xuca1 |
|
not modelled |
44.8 |
42 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 33 | d1hova_ |
|
not modelled |
44.6 |
19 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 34 | d1rm8a_ |
|
not modelled |
44.6 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 35 | d2ovxa1 |
|
not modelled |
44.4 |
36 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Matrix metalloproteases, catalytic domain |
| 36 | c1su3A_ |
|
not modelled |
41.5 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:interstitial collagenase;
PDBTitle: x-ray structure of human prommp-1: new insights into2 collagenase action
|
| 37 | d1sata2 |
|
not modelled |
41.3 |
17 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Serralysin-like metalloprotease, catalytic (N-terminal) domain |
| 38 | c3ba0A_ |
|
not modelled |
40.3 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:macrophage metalloelastase;
PDBTitle: crystal structure of full-length human mmp-12
|
| 39 | c3b4rB_ |
|
not modelled |
40.2 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative zinc metalloprotease mj0392;
PDBTitle: site-2 protease from methanocaldococcus jannaschii
|
| 40 | c1slmA_ |
|
not modelled |
39.7 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:stromelysin-1;
PDBTitle: crystal structure of fibroblast stromelysin-1: the c-truncated human2 proenzyme
|
| 41 | d2ixna1 |
|
not modelled |
36.7 |
16 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 42 | c1jiwP_ |
|
not modelled |
36.3 |
22 |
PDB header:hydrolase/hyrolase inhibitor
Chain: P: PDB Molecule:alkaline metalloproteinase;
PDBTitle: crystal structure of the apr-aprin complex
|
| 43 | c3d7iB_ |
|
not modelled |
35.2 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:carboxymuconolactone decarboxylase family protein;
PDBTitle: crystal structure of carboxymuconolactone decarboxylase family protein2 possibly involved in oxygen detoxification (1591455) from3 methanococcus jannaschii at 1.75 a resolution
|
| 44 | d1kjpa_ |
|
not modelled |
34.6 |
27 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 45 | d1lmla_ |
|
not modelled |
33.9 |
42 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Leishmanolysin |
| 46 | c2cltB_ |
|
not modelled |
33.8 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:interstitial collagenase;
PDBTitle: crystal structure of the active form (full-length) of human2 fibroblast collagenase.
|
| 47 | d1bqba_ |
|
not modelled |
33.3 |
18 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 48 | c1y791_ |
|
not modelled |
32.8 |
27 |
PDB header:hydrolase
Chain: 1: PDB Molecule:peptidyl-dipeptidase dcp;
PDBTitle: crystal structure of the e.coli dipeptidyl carboxypeptidase2 dcp in complex with a peptidic inhibitor
|
| 49 | c1eakA_ |
|
not modelled |
32.6 |
10 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:72 kda type iv collagenase;
PDBTitle: catalytic domain of prommp-2 e404q mutant
|
| 50 | d3e11a1 |
|
not modelled |
29.3 |
33 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TTHA0227-like |
| 51 | c3b7uX_ |
|
not modelled |
29.0 |
32 |
PDB header:hydrolase
Chain: X: PDB Molecule:leukotriene a-4 hydrolase;
PDBTitle: leukotriene a4 hydrolase complexed with kelatorphan
|
| 52 | c1l6jA_ |
|
not modelled |
23.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:matrix metalloproteinase-9;
PDBTitle: crystal structure of human matrix metalloproteinase mmp92 (gelatinase b).
|
| 53 | c1om8A_ |
|
not modelled |
23.6 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:serralysin;
PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
|
| 54 | d1neka3 |
|
not modelled |
23.1 |
24 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 55 | c2j83B_ |
|
not modelled |
22.5 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:ulilysin;
PDBTitle: ulilysin metalloprotease in complex with batimastat.
|
| 56 | c3c0hB_ |
|
not modelled |
21.4 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:peripheral plasma membrane protein cask;
PDBTitle: cask cam-kinase domain- amppnp complex, p1 form
|
| 57 | c1satA_ |
|
not modelled |
20.8 |
17 |
PDB header:hydrolase (serine protease)
Chain: A: PDB Molecule:serratia protease;
PDBTitle: crystal structure of the 50 kda metallo protease from s.2 marcescens
|
| 58 | d3b7sa3 |
|
not modelled |
20.0 |
40 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Leukotriene A4 hydrolase catalytic domain |
| 59 | c2oqjL_ |
|
not modelled |
19.0 |
50 |
PDB header:immune system
Chain: L: PDB Molecule:peptide 2g12.1 (acppshvldmrsgtclaaegk);
PDBTitle: crystal structure analysis of fab 2g12 in complex with peptide 2g12.1
|
| 60 | c2oqjC_ |
|
not modelled |
18.9 |
50 |
PDB header:immune system
Chain: C: PDB Molecule:peptide 2g12.1 (acppshvldmrsgtclaaegk);
PDBTitle: crystal structure analysis of fab 2g12 in complex with peptide 2g12.1
|
| 61 | d1kufa_ |
|
not modelled |
18.8 |
23 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 62 | c2oqjI_ |
|
not modelled |
18.7 |
50 |
PDB header:immune system
Chain: I: PDB Molecule:peptide 2g12.1 (acppshvldmrsgtclaaegk);
PDBTitle: crystal structure analysis of fab 2g12 in complex with peptide 2g12.1
|
| 63 | c3k7lA_ |
|
not modelled |
17.9 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:atragin;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 64 | d2ixma1 |
|
not modelled |
17.8 |
18 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 65 | c2gtqA_ |
|
not modelled |
17.7 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:aminopeptidase n;
PDBTitle: crystal structure of aminopeptidase n from human pathogen neisseria2 meningitidis
|
| 66 | d1npca_ |
|
not modelled |
16.7 |
20 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 67 | c2g62A_ |
|
not modelled |
16.4 |
22 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:protein phosphatase 2a, regulatory subunit b' (pr 53);
PDBTitle: crystal structure of human ptpa
|
| 68 | c2oqjF_ |
|
not modelled |
15.7 |
50 |
PDB header:immune system
Chain: F: PDB Molecule:peptide 2g12.1 (acppshvldmrsgtclaaegk);
PDBTitle: crystal structure analysis of fab 2g12 in complex with peptide 2g12.1
|
| 69 | c2erpA_ |
|
not modelled |
15.4 |
29 |
PDB header:toxin
Chain: A: PDB Molecule:vascular apoptosis-inducing protein 1;
PDBTitle: crystal structure of vascular apoptosis-inducing protein-1(inhibitor-2 bound form)
|
| 70 | c3k7nA_ |
|
not modelled |
15.3 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:k-like;
PDBTitle: structures of two elapid snake venom metalloproteases with2 distinct activities highlight the disulfide patterns in the3 d domain of adamalysin family proteins
|
| 71 | c3ebhA_ |
|
not modelled |
14.9 |
30 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:m1 family aminopeptidase;
PDBTitle: structure of the m1 alanylaminopeptidase from malaria complexed with2 bestatin
|
| 72 | d1wlfa2 |
|
not modelled |
14.8 |
27 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Cdc48 N-terminal domain-like |
| 73 | c2c1iA_ |
|
not modelled |
14.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan glcnac deacetylase;
PDBTitle: structure of streptococcus pnemoniae peptidoglycan2 deacetylase (sppgda) d 275 n mutant.
|
| 74 | c2i47A_ |
|
not modelled |
14.7 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:adam 17;
PDBTitle: crystal structure of catalytic domain of tace with inhibitor
|
| 75 | c1wgzC_ |
|
not modelled |
14.5 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:carboxypeptidase 1;
PDBTitle: crystal structure of carboxypeptidase 1 from thermus thermophilus
|
| 76 | c2c30A_ |
|
not modelled |
14.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine-protein kinase pak 6;
PDBTitle: crystal structure of the human p21-activated kinase 6
|
| 77 | c3b37A_ |
|
not modelled |
13.9 |
40 |
PDB header:hydrolase
Chain: A: PDB Molecule:aminopeptidase n;
PDBTitle: crystal structure of e. coli aminopeptidase n in complex with tyrosine
|
| 78 | d2i47a1 |
|
not modelled |
13.6 |
21 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:TNF-alpha converting enzyme, TACE, catalytic domain |
| 79 | c3ppuB_ |
|
not modelled |
13.2 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione-s-transferase;
PDBTitle: crystal structure of the glutathione-s-transferase xi from2 phanerochaete chrysosporium
|
| 80 | c1yp1A_ |
|
not modelled |
13.1 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:fii;
PDBTitle: crystal structure of a non-hemorrhagic fibrin(ogen)olytic2 metalloproteinase from venom of agkistrodon acutus
|
| 81 | d2ajfa1 |
|
not modelled |
12.9 |
11 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Neurolysin-like |
| 82 | d1nd1a_ |
|
not modelled |
12.9 |
29 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 83 | c3famA_ |
|
not modelled |
12.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:carbon catabolite-derepressing protein kinase;
PDBTitle: crystal structure of the protein kinase domain of yeast amp-2 activated protein kinase snf1
|
| 84 | c1yyeA_ |
|
not modelled |
12.8 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:estrogen receptor beta;
PDBTitle: crystal structure of estrogen receptor beta complexed with2 way-202196
|
| 85 | d1eb6a_ |
|
not modelled |
12.5 |
16 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Fungal zinc peptidase |
| 86 | c1z5hB_ |
|
not modelled |
12.4 |
60 |
PDB header:hydrolase
Chain: B: PDB Molecule:tricorn protease interacting factor f3;
PDBTitle: crystal structures of the tricorn interacting factor f32 from thermoplasma acidophilum
|
| 87 | c2v4bB_ |
|
not modelled |
12.3 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:adamts-1;
PDBTitle: crystal structure of human adamts-1 catalytic domain and2 cysteine-rich domain (apo-form)
|
| 88 | c3cqbB_ |
|
not modelled |
12.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable protease htpx homolog;
PDBTitle: crystal structure of heat shock protein htpx domain from vibrio2 parahaemolyticus rimd 2210633
|
| 89 | d1phka_ |
|
not modelled |
12.2 |
9 |
Fold:Protein kinase-like (PK-like)
Superfamily:Protein kinase-like (PK-like)
Family:Protein kinases, catalytic subunit |
| 90 | c2e3xA_ |
|
not modelled |
12.2 |
29 |
PDB header:hydrolase, blood clotting, toxin
Chain: A: PDB Molecule:coagulation factor x-activating enzyme heavy chain;
PDBTitle: crystal structure of russell's viper venom metalloproteinase
|
| 91 | c3g5cA_ |
|
not modelled |
12.1 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:adam 22;
PDBTitle: structural and biochemical studies on the ectodomain of human adam22
|
| 92 | c3nqzB_ |
|
not modelled |
11.9 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:secreted metalloprotease mcp02;
PDBTitle: crystal structure of the autoprocessed vibriolysin mcp-02 with e369a2 mutation
|
| 93 | d1atla_ |
|
not modelled |
11.9 |
29 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Reprolysin-like |
| 94 | c3b8zB_ |
|
not modelled |
11.8 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein adamts-5;
PDBTitle: high resolution crystal structure of the catalytic domain2 of adamts-5 (aggrecanase-2)
|
| 95 | d1chua3 |
|
not modelled |
11.8 |
14 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 96 | c3ce2A_ |
|
not modelled |
11.7 |
36 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative peptidase;
PDBTitle: crystal structure of putative peptidase from chlamydophila abortus
|
| 97 | c2rjpC_ |
|
not modelled |
11.5 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:adamts-4;
PDBTitle: crystal structure of adamts4 with inhibitor bound
|
| 98 | c2rjqA_ |
|
not modelled |
11.3 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:adamts-5;
PDBTitle: crystal structure of adamts5 with inhibitor bound
|
| 99 | c2dw1B_ |
|
not modelled |
11.3 |
29 |
PDB header:apoptosis, toxin
Chain: B: PDB Molecule:catrocollastatin;
PDBTitle: crystal structure of vap2 from crotalus atrox venom (form 2-2 crystal)
|