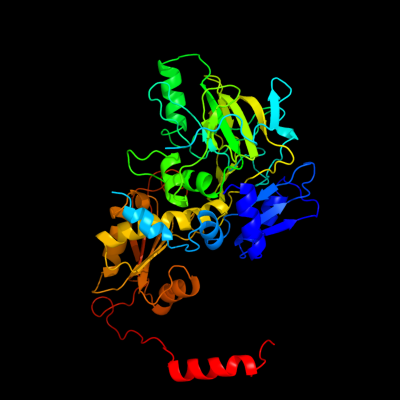
| 1 | c2idbB_
|
|
 |
100.0 |
98 |
PDB header:lyase
Chain: B: PDB Molecule:3-octaprenyl-4-hydroxybenzoate carboxy-lyase;
PDBTitle: crystal structure of 3-octaprenyl-4-hydroxybenzoate decarboxylase2 (ubid) from escherichia coli, northeast structural genomics target3 er459.
|
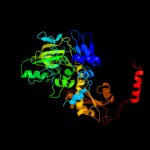
| 2 | d2idba1
|
|
 |
100.0 |
98 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:UbiD middle domain-like |
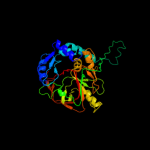
| 3 | d2idba2
|
|
 |
100.0 |
97 |
Fold:UbiD C-terminal domain-like
Superfamily:UbiD C-terminal domain-like
Family:UbiD C-terminal domain-like |
| 4 | c3bpkB_
|
|
 |
68.5 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase component b;
PDBTitle: crystal structure of nitrilotriacetate monooxygenase2 component b from bacillus cereus
|
| 5 | c3e4vA_
|
|
 |
59.7 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:nadh:fmn oxidoreductase like protein;
PDBTitle: crystal structure of nadh:fmn oxidoreductase like protein in complex2 with fmn (yp_544701.1) from methylobacillus flagellatus kt at 1.40 a3 resolution
|
| 6 | d1ejea_
|
|
 |
57.5 |
14 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 7 | c3ibpA_
|
|
 |
35.8 |
22 |
PDB header:cell cycle
Chain: A: PDB Molecule:chromosome partition protein mukb;
PDBTitle: the crystal structure of the dimerization domain of escherichia coli2 structural maintenance of chromosomes protein mukb
|
| 8 | c3fgeA_
|
|
 |
17.1 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative flavin reductase with split barrel domain;
PDBTitle: crystal structure of putative flavin reductase with split barrel2 domain (yp_750721.1) from shewanella frigidimarina ncimb 400 at 1.743 a resolution
|
| 9 | c2k29A_
|
|
 |
16.8 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
| 10 | c2iu6B_
|
|
 |
16.6 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:dihydroxyacetone kinase;
PDBTitle: regulation of the dha operon of lactococcus lactis
|
| 11 | c2i46A_
|
|
 |
16.5 |
23 |
PDB header:protein binding
Chain: A: PDB Molecule:adrenocortical dysplasia protein homolog;
PDBTitle: crystal structure of human tpp1
|
| 12 | d1j98a_
|
|
 |
16.4 |
67 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 13 | c1uv7A_
|
|
 |
15.8 |
10 |
PDB header:transport
Chain: A: PDB Molecule:general secretion pathway protein m;
PDBTitle: periplasmic domain of epsm from vibrio cholerae
|
| 14 | d1uv7a_
|
|
 |
15.8 |
10 |
Fold:RRF/tRNA synthetase additional domain-like
Superfamily:General secretion pathway protein M, EpsM
Family:General secretion pathway protein M, EpsM |
| 15 | d1oi2a_
|
|
 |
15.7 |
26 |
Fold:DAK1/DegV-like
Superfamily:DAK1/DegV-like
Family:DAK1 |
| 16 | c3ct4B_
|
|
 |
15.1 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:pts-dependent dihydroxyacetone kinase,
PDBTitle: structure of dha-kinase subunit dhak from l. lactis
|
| 17 | d1vjea_
|
|
 |
15.1 |
56 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 18 | c2pjpA_
|
|
 |
14.5 |
21 |
PDB header:translation/rna
Chain: A: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: structure of the mrna-binding domain of elongation factor2 selb from e.coli in complex with secis rna
|
| 19 | d1lvaa3
|
|
 |
14.5 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 20 | d1un8a4
|
|
 |
14.2 |
19 |
Fold:DAK1/DegV-like
Superfamily:DAK1/DegV-like
Family:DAK1 |
| 21 | d1h32a1 |
|
not modelled |
13.1 |
18 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c SoxA |
| 22 | d2bj7a1 |
|
not modelled |
12.5 |
19 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 23 | d2ezwa1 |
|
not modelled |
12.4 |
28 |
Fold:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Superfamily:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Family:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit |
| 24 | c1iweB_ |
|
not modelled |
12.1 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: imp complex of the recombinant mouse-muscle2 adenylosuccinate synthetase
|
| 25 | d1cf1a1 |
|
not modelled |
10.7 |
11 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arrestin/Vps26-like |
| 26 | c2ixaA_ |
|
not modelled |
10.4 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 27 | d1iwea_ |
|
not modelled |
9.5 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 28 | c2b7fD_ |
|
not modelled |
9.4 |
23 |
PDB header:hydrolase/hydrolase inhibitor
Chain: D: PDB Molecule:htlv protease;
PDBTitle: crystal structure of human t-cell leukemia virus protease, a novel2 target for anti-cancer design
|
| 29 | c2wmmA_ |
|
not modelled |
9.2 |
25 |
PDB header:cell cycle
Chain: A: PDB Molecule:chromosome partition protein mukb;
PDBTitle: crystal structure of the hinge domain of mukb
|
| 30 | d1p9ba_ |
|
not modelled |
9.2 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 31 | c2rjzA_ |
|
not modelled |
9.1 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pilo protein;
PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
|
| 32 | c3nuhB_ |
|
not modelled |
9.1 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit b;
PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
|
| 33 | c3ip3D_ |
|
not modelled |
8.3 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, putative;
PDBTitle: structure of putative oxidoreductase (tm_0425) from2 thermotoga maritima
|
| 34 | c1un9B_ |
|
not modelled |
8.1 |
19 |
PDB header:kinase
Chain: B: PDB Molecule:dihydroxyacetone kinase;
PDBTitle: crystal structure of the dihydroxyacetone kinase from c.2 freundii in complex with amp-pnp and mg2+
|
| 35 | d1ldda_ |
|
not modelled |
7.8 |
35 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:SCF ubiquitin ligase complex WHB domain |
| 36 | d1kwga1 |
|
not modelled |
7.8 |
13 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 37 | c5acnA_ |
|
not modelled |
7.0 |
18 |
PDB header:lyase(carbon-oxygen)
Chain: A: PDB Molecule:aconitase;
PDBTitle: structure of activated aconitase. formation of the (4fe-4s)2 cluster in the crystal
|
| 38 | d1acoa2 |
|
not modelled |
7.0 |
18 |
Fold:Aconitase iron-sulfur domain
Superfamily:Aconitase iron-sulfur domain
Family:Aconitase iron-sulfur domain |
| 39 | d1g8fa3 |
|
not modelled |
6.9 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ATP sulfurylase C-terminal domain |
| 40 | c1ayrA_ |
|
not modelled |
6.8 |
12 |
PDB header:sensory transduction
Chain: A: PDB Molecule:arrestin;
PDBTitle: arrestin from bovine rod outer segments
|
| 41 | d1vjja3 |
|
not modelled |
6.8 |
24 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 42 | d2gaxa1 |
|
not modelled |
6.5 |
31 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 43 | c2zjtB_ |
|
not modelled |
6.4 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit b;
PDBTitle: crystal structure of dna gyrase b' domain sheds lights on2 the mechanism for t-segment navigation
|
| 44 | d1qmga1 |
|
not modelled |
6.2 |
43 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:Acetohydroxy acid isomeroreductase (ketol-acid reductoisomerase, KARI) |
| 45 | d1na6a1 |
|
not modelled |
5.9 |
33 |
Fold:DNA-binding pseudobarrel domain
Superfamily:DNA-binding pseudobarrel domain
Family:Type II restriction endonuclease effector domain |
| 46 | d3bvua3 |
|
not modelled |
5.9 |
19 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:alpha-mannosidase |
| 47 | d1ex0a3 |
|
not modelled |
5.9 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 48 | c2ow7A_ |
|
not modelled |
5.9 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase 2;
PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
|
| 49 | c3fofD_ |
|
not modelled |
5.8 |
11 |
PDB header:isomerase/dna
Chain: D: PDB Molecule:dna topoisomerase 4 subunit b;
PDBTitle: structural insight into the quinolone-dna cleavage complex2 of type iia topoisomerases
|
| 50 | d2hzab1 |
|
not modelled |
5.7 |
19 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 51 | c3bnkB_ |
|
not modelled |
5.7 |
14 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoredoxin;
PDBTitle: x-ray crystal structure of flavoredoxin from methanosarcina2 acetivorans
|
| 52 | c1htyA_ |
|
not modelled |
5.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase ii;
PDBTitle: golgi alpha-mannosidase ii
|
| 53 | c2zkra_ |
|
not modelled |
5.6 |
18 |
PDB header:ribosomal protein/rna
Chain: A: PDB Molecule:rna expansion segment es3;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 54 | d1rl6a1 |
|
not modelled |
5.4 |
30 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 55 | d1vqoe1 |
|
not modelled |
5.3 |
26 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 56 | c3r7tA_ |
|
not modelled |
5.1 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of adenylosuccinate synthetase from campylobacter2 jejuni
|