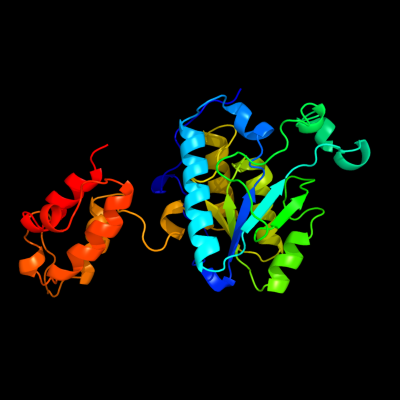
| 1 | c2fq1A_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:isochorismatase;
PDBTitle: crystal structure of the two-domain non-ribosomal peptide synthetase2 entb containing isochorismate lyase and aryl-carrier protein domains
|
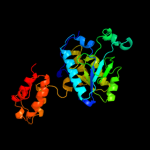
| 2 | d1nf9a_
|
|
 |
100.0 |
47 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
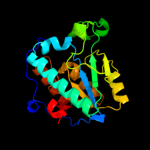
| 3 | d1nbaa_
|
|
 |
100.0 |
23 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
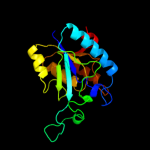
| 4 | c3irvA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:cysteine hydrolase;
PDBTitle: crystal structure of cysteine hydrolase pspph_2384 from pseudomonas2 syringae pv. phaseolicola 1448a
|
| 5 | c3ot4F_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 6 | d1j2ra_
|
|
 |
100.0 |
21 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 7 | c3kl2K_
|
|
 |
100.0 |
20 |
PDB header:structural genomics, unknown function
Chain: K: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase from streptomyces2 avermitilis
|
| 8 | c3oqpA_
|
|
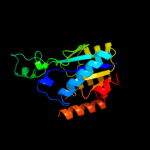 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 9 | c3oqpB_
|
|
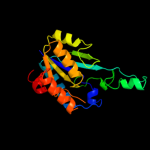 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 10 | c3hu5B_
|
|
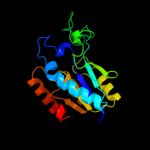 |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 11 | c3mcwA_
|
|
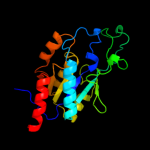 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of an a putative hydrolase of the isochorismatase2 family (cv_1320) from chromobacterium violaceum atcc 12472 at 1.06 a3 resolution
|
| 12 | c3hb7G_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: G: PDB Molecule:isochorismatase hydrolase;
PDBTitle: the crystal structure of an isochorismatase-like hydrolase from2 alkaliphilus metalliredigens to 2.3a
|
| 13 | c3eefA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-carbamoylsarcosine amidase related protein;
PDBTitle: crystal structure of n-carbamoylsarcosine amidase from thermoplasma2 acidophilum
|
| 14 | d1im5a_
|
|
 |
100.0 |
20 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 15 | c3r2jC_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
| 16 | c3lqyA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase hydrolase;
PDBTitle: crystal structure of putative isochorismatase hydrolase from2 oleispira antarctica
|
| 17 | c3o93A_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
|
| 18 | c2wtaA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
|
| 19 | c3gbcA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 20 | d1yaca_
|
|
 |
100.0 |
20 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 21 | c2a67C_ |
|
not modelled |
100.0 |
20 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein
|
| 22 | c2b34C_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 23 | c1yzvA_ |
|
not modelled |
100.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
| 24 | d1x9ga_ |
|
not modelled |
100.0 |
16 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 25 | c2h0rD_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
| 26 | c2cq8A_ |
|
not modelled |
99.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: solution structure of rsgi ruh-033, a pp-binding domain of2 10-fthfdh from human cdna
|
| 27 | c2jgpA_ |
|
not modelled |
99.6 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:tyrocidine synthetase 3;
PDBTitle: structure of the tycc5-6 pcp-c bidomain of the tyrocidine2 synthetase tycc
|
| 28 | c2roqA_ |
|
not modelled |
99.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:enterobactin synthetase component f;
PDBTitle: solution structure of the thiolation-thioesterase di-domain2 of enterobactin synthetase component f
|
| 29 | c2vsqA_ |
|
not modelled |
99.6 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:surfactin synthetase subunit 3;
PDBTitle: structure of surfactin a synthetase c (srfa-c), a2 nonribosomal peptide synthetase termination module
|
| 30 | d2gdwa1 |
|
not modelled |
99.5 |
27 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Peptidyl carrier domain |
| 31 | c2ju2A_ |
|
not modelled |
99.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:erythronolide synthase;
PDBTitle: minimized mean solution structure of the acyl carrier2 protein domain from module 2 of 6-deoxyerythronolide b3 synthase (debs)
|
| 32 | c2liuA_ |
|
not modelled |
99.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:cura;
PDBTitle: nmr structure of holo-acpi domain from cura module from lyngbya2 majuscula
|
| 33 | c2afdA_ |
|
not modelled |
99.5 |
22 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:protein asl1650;
PDBTitle: solution structure of asl1650, an acyl carrier protein from anabaena2 sp. pcc 7120 with a variant phosphopantetheinylation-site sequence
|
| 34 | c2kr5A_ |
|
not modelled |
99.3 |
21 |
PDB header:transport protein
Chain: A: PDB Molecule:aflatoxin biosynthesis polyketide synthase;
PDBTitle: solution structure of an acyl carrier protein domain from fungal type2 i polyketide synthase
|
| 35 | d2af8a_ |
|
not modelled |
99.3 |
22 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 36 | d1or5a_ |
|
not modelled |
99.3 |
15 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 37 | c2ehtA_ |
|
not modelled |
99.3 |
19 |
PDB header:lipid transport
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: crystal structure of acyl carrier protein from aquifex aeolicus (form2 2)
|
| 38 | d1nq4a_ |
|
not modelled |
99.3 |
17 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 39 | d1vkua_ |
|
not modelled |
99.3 |
10 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 40 | d1klpa_ |
|
not modelled |
99.3 |
21 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 41 | d1t8ka_ |
|
not modelled |
99.2 |
17 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 42 | c1x3oA_ |
|
not modelled |
99.2 |
16 |
PDB header:lipid transport
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: crystal structure of the acyl carrier protein from thermus2 thermophilus hb8
|
| 43 | c2fq2A_ |
|
not modelled |
99.2 |
17 |
PDB header:lipid transport
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of minor conformation of holo-acyl2 carrier protein from malaria parasite plasmodium falciparum
|
| 44 | c2qnwA_ |
|
not modelled |
99.2 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: toxoplasma gondii apicoplast-targeted acyl carrier protein
|
| 45 | c2cgqA_ |
|
not modelled |
99.2 |
17 |
PDB header:protein transport
Chain: A: PDB Molecule:acyl carrier protein acpa;
PDBTitle: a putative acyl carrier protein(rv0033) from mycobacterium2 tuberculosis
|
| 46 | c2l3vA_ |
|
not modelled |
99.2 |
15 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: nmr structure of acyl carrier protein from brucella melitensis
|
| 47 | d1f80d_ |
|
not modelled |
99.2 |
9 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 48 | c2cnrA_ |
|
not modelled |
99.1 |
17 |
PDB header:lipid transport
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: structural studies on the interaction of scfas acp with2 acps
|
| 49 | d2pnga1 |
|
not modelled |
99.1 |
15 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 50 | c3ejbC_ |
|
not modelled |
99.1 |
17 |
PDB header:oxidoreductase/lipid transport
Chain: C: PDB Molecule:acyl carrier protein;
PDBTitle: crystal structure of p450bioi in complex with tetradecanoic2 acid ligated acyl carrier protein
|
| 51 | c2fvfA_ |
|
not modelled |
99.1 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: structure of 10:0-acp (protein with docked fatty acid)
|
| 52 | c2jq4A_ |
|
not modelled |
99.1 |
20 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical protein atu2571;
PDBTitle: complete resonance assignments and solution structure2 calculation of atc2521 (nesg id: att6) from agrobacterium3 tumefaciens
|
| 53 | d2jq4a1 |
|
not modelled |
99.1 |
20 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 54 | c2l9fA_ |
|
not modelled |
99.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:cale8;
PDBTitle: nmr solution structure of meacp
|
| 55 | c2dnwA_ |
|
not modelled |
99.0 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of rsgi ruh-059, an acp domain of acyl2 carrier protein, mitochondrial [precursor] from human cdna
|
| 56 | c2kciA_ |
|
not modelled |
99.0 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative acyl carrier protein;
PDBTitle: solution nmr structure of gmet_2339 from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr141
|
| 57 | c2kwlA_ |
|
not modelled |
99.0 |
18 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of acyl carrier protein from borrelia burgdorferi
|
| 58 | c2l4bA_ |
|
not modelled |
99.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of a putative acyl carrier protein from anaplasma2 phagocytophilum. seattle structural genomics center for infectious3 disease target anpha.01018.a
|
| 59 | d1dv5a_ |
|
not modelled |
98.9 |
19 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:apo-D-alanyl carrier protein |
| 60 | c3lmoA_ |
|
not modelled |
98.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:specialized acyl carrier protein;
PDBTitle: crystal structure of specialized acyl carrier protein2 (rpa2022) from rhodopseudomonas palustris, northeast3 structural genomics consortium target rpr324
|
| 61 | c3ce7A_ |
|
not modelled |
98.6 |
10 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:specific mitochodrial acyl carrier protein;
PDBTitle: crystal structure of toxoplasma specific mitochodrial acyl2 carrier protein, 59.m03510
|
| 62 | c2vkzC_ |
|
not modelled |
95.4 |
5 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 63 | c2uv8C_ |
|
not modelled |
94.7 |
5 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha (fas2);
PDBTitle: crystal structure of yeast fatty acid synthase with stalled2 acyl carrier protein at 3.1 angstrom resolution
|
| 64 | c3s8mA_ |
|
not modelled |
92.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acp reductase;
PDBTitle: the crystal structure of fabv
|
| 65 | c2uv9B_ |
|
not modelled |
89.1 |
4 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase alpha subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the alpha subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 66 | d1xp8a1 |
|
not modelled |
77.0 |
6 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 67 | d1cmwa2 |
|
not modelled |
76.6 |
17 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 68 | d1f2da_ |
|
not modelled |
71.9 |
9 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 69 | c1x1qA_ |
|
not modelled |
66.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
|
| 70 | d2zdra2 |
|
not modelled |
65.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 71 | c3bgwD_ |
|
not modelled |
63.6 |
14 |
PDB header:replication
Chain: D: PDB Molecule:dnab-like replicative helicase;
PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
|
| 72 | c1xuzA_ |
|
not modelled |
62.9 |
10 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 73 | c2iusB_ |
|
not modelled |
62.6 |
15 |
PDB header:membrane protein
Chain: B: PDB Molecule:dna translocase ftsk;
PDBTitle: e. coli ftsk motor domain
|
| 74 | c2iutA_ |
|
not modelled |
59.0 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:dna translocase ftsk;
PDBTitle: p. aeruginosa ftsk motor domain, dimeric
|
| 75 | c2iuuE_ |
|
not modelled |
58.6 |
15 |
PDB header:membrane protein
Chain: E: PDB Molecule:dna translocase ftsk;
PDBTitle: p. aeruginosa ftsk motor domain, hexamer
|
| 76 | c2recB_ |
|
not modelled |
58.0 |
11 |
PDB header:helicase
PDB COMPND:
|
| 77 | d1ubea1 |
|
not modelled |
56.7 |
6 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 78 | d1vp8a_ |
|
not modelled |
56.2 |
19 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 79 | c3g8rA_ |
|
not modelled |
55.7 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 80 | d1vlia2 |
|
not modelled |
55.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 81 | c2vyeA_ |
|
not modelled |
53.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the dnac-ssdna complex
|
| 82 | c1xp8A_ |
|
not modelled |
53.0 |
6 |
PDB header:dna binding protein
Chain: A: PDB Molecule:reca protein;
PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
|
| 83 | c3cbwA_ |
|
not modelled |
48.4 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ydht protein;
PDBTitle: crystal structure of the ydht protein from bacillus subtilis
|
| 84 | c2ihnA_ |
|
not modelled |
45.9 |
7 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:ribonuclease h;
PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
|
| 85 | d1xu9a_ |
|
not modelled |
44.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 86 | d2ae2a_ |
|
not modelled |
44.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 87 | c3nugA_ |
|
not modelled |
44.2 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier protein) reductase;
PDBTitle: crystal structure of wild type tetrameric pyridoxal 4-dehydrogenase2 from mesorhizobium loti
|
| 88 | d1tfra2 |
|
not modelled |
42.3 |
7 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 89 | c3ijrF_ |
|
not modelled |
42.2 |
17 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
| 90 | d1ae1a_ |
|
not modelled |
41.8 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 91 | d1gz6a_ |
|
not modelled |
41.7 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 92 | d1xsea_ |
|
not modelled |
41.2 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 93 | c2cvhB_ |
|
not modelled |
40.4 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dna repair and recombination protein radb;
PDBTitle: crystal structure of the radb recombinase
|
| 94 | d2a4ka1 |
|
not modelled |
40.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 95 | c2pd6D_ |
|
not modelled |
40.2 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:estradiol 17-beta-dehydrogenase 8;
PDBTitle: structure of human hydroxysteroid dehydrogenase type 8, hsd17b8
|
| 96 | c3ai3A_ |
|
not modelled |
40.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph-sorbose reductase;
PDBTitle: the crystal structure of l-sorbose reductase from gluconobacter2 frateurii complexed with nadph and l-sorbose
|
| 97 | c1vliA_ |
|
not modelled |
39.5 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 98 | d1mo6a1 |
|
not modelled |
37.2 |
6 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 99 | d1e0ta2 |
|
not modelled |
37.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 100 | c3bh0A_ |
|
not modelled |
37.0 |
13 |
PDB header:replication
Chain: A: PDB Molecule:dnab-like replicative helicase;
PDBTitle: atpase domain of g40p
|
| 101 | c2nwqA_ |
|
not modelled |
36.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable short-chain dehydrogenase;
PDBTitle: short chain dehydrogenase from pseudomonas aeruginosa
|
| 102 | c2y8kA_ |
|
not modelled |
35.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate binding family 6;
PDBTitle: structure of ctgh5-cbm6, an arabinoxylan-specific xylanase.
|
| 103 | c2nm0B_ |
|
not modelled |
35.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable 3-oxacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of sco1815: a beta-ketoacyl-acyl carrier protein2 reductase from streptomyces coelicolor a3(2)
|
| 104 | c3ldaA_ |
|
not modelled |
35.6 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna repair protein rad51;
PDBTitle: yeast rad51 h352y filament interface mutant
|
| 105 | c3toxG_ |
|
not modelled |
34.3 |
21 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 106 | c3n7tA_ |
|
not modelled |
33.0 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:macrophage binding protein;
PDBTitle: crystal structure of a macrophage binding protein from coccidioides2 immitis
|
| 107 | d1pkla2 |
|
not modelled |
32.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 108 | c2zroA_ |
|
not modelled |
32.1 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein reca;
PDBTitle: msreca adp form iv
|
| 109 | c3o26A_ |
|
not modelled |
31.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:salutaridine reductase;
PDBTitle: the structure of salutaridine reductase from papaver somniferum.
|
| 110 | d1wdea_ |
|
not modelled |
31.3 |
15 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 111 | c3enoB_ |
|
not modelled |
30.1 |
21 |
PDB header:hydrolase/unknown function
Chain: B: PDB Molecule:putative o-sialoglycoprotein endopeptidase;
PDBTitle: crystal structure of pyrococcus furiosus pcc1 in complex2 with thermoplasma acidophilum kae1
|
| 112 | d1a3xa2 |
|
not modelled |
30.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 113 | c2dflA_ |
|
not modelled |
29.9 |
15 |
PDB header:recombination
Chain: A: PDB Molecule:dna repair and recombination protein rada;
PDBTitle: crystal structure of left-handed rada filament
|
| 114 | c2b4qB_ |
|
not modelled |
28.8 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rhamnolipids biosynthesis 3-oxoacyl-[acyl-
PDBTitle: pseudomonas aeruginosa rhlg/nadp active-site complex
|
| 115 | c2amwA_ |
|
not modelled |
28.4 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ne2163;
PDBTitle: solution nmr structure of protein ne2163 from nitrosomonas europaea.2 northeast structural genomics consortium target net1.
|
| 116 | c2lkiA_ |
|
not modelled |
28.4 |
13 |
PDB header:lipid transport
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution nmr structure of holo acyl carrier protein ne2163 from2 nitrosomonas europaea. northeast structural genomics consortium3 target net1.
|
| 117 | d1o4wa_ |
|
not modelled |
28.2 |
37 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:PIN domain |
| 118 | d1qopb_ |
|
not modelled |
28.2 |
13 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 119 | c3sjuA_ |
|
not modelled |
28.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:keto reductase;
PDBTitle: hedamycin polyketide ketoreductase bound to nadph
|
| 120 | d2pd4a1 |
|
not modelled |
27.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |