1 d1df6a_
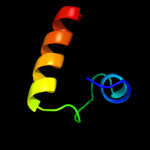
61.5
100
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Cycloviolacin2 c1df6A_
61.5
100
PDB header: plant proteinChain: A: PDB Molecule: cycloviolacin o1;PDBTitle: 1h nmr solution structure of cycloviolacin o1
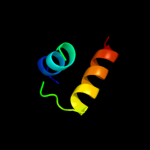
3 d2bjca1
48.6
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator4 d1lcda_
48.5
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator5 d1qpza1
47.0
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator6 c2l8nA_
45.2
23
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional repressor cytr;PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
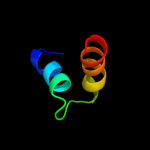
7 d1nbja_
43.9
100
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Cycloviolacin8 c1nbjA_
43.9
100
PDB header: plant proteinChain: A: PDB Molecule: cycloviolacin o1;PDBTitle: high-resolution solution structure of cycloviolacin o1
9 c2c3hC_
41.0
36
PDB header: carbohydrate-binding moduleChain: C: PDB Molecule: alpha-amylase g-6;PDBTitle: structure of cbm26 from bacillus halodurans amylase in2 complex with maltose
10 d1uxca_
38.6
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator11 c2b5kA_
37.8
55
PDB header: antimicrobial proteinChain: A: PDB Molecule: polyphemusin-1;PDBTitle: pv5 nmr solution structure in dpc micelles
12 d2phcb1
36.4
36
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like13 c2lcvA_
31.9
23
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
14 d1efaa1
29.6
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator15 c3bd1B_
24.4
14
PDB header: transcriptionChain: B: PDB Molecule: cro protein;PDBTitle: structure of the cro protein from putative prophage element xfaso 1 in2 xylella fastidiosa strain ann-1
16 c2zp2B_
22.1
42
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
17 c2vt1A_
22.0
22
PDB header: membrane proteinChain: A: PDB Molecule: surface presentation of antigens protein spas;PDBTitle: crystal structure of the cytoplasmic domain of spa40, the2 specificity switch for the shigella flexneri type iii3 secretion system
18 c3h9xB_
20.8
28
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein pspto_3016;PDBTitle: crystal structure of the pspto_3016 protein from2 pseudomonas syringae, northeast structural genomics3 consortium target psr293
19 d1uxda_
19.6
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator20 c3hgtA_
18.5
29
PDB header: transcriptionChain: A: PDB Molecule: hda1 complex subunit 3;PDBTitle: structural and functional studies of the yeast class ii hda12 hdac complex
21 c2phcB_
not modelled
16.6
33
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
22 d2ptda_
not modelled
16.4
10
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Bacterial PLC23 d2duca1
not modelled
16.2
37
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold24 d2hsga1
not modelled
16.0
29
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator25 d2elca1
not modelled
15.5
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain26 c2rp4C_
not modelled
15.5
20
PDB header: transcriptionChain: C: PDB Molecule: transcription factor p53;PDBTitle: solution structure of the oligomerization domain in dmp53
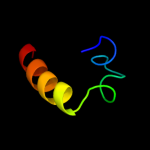
27 d1vb8a_
not modelled
15.0
100
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Cycloviolacin28 c1vb8A_
not modelled
15.0
100
PDB header: plant proteinChain: A: PDB Molecule: viola hederacea root peptide 1;PDBTitle: solution structure of vhr1, the first cyclotide from root2 tissue
29 c1bdhA_
not modelled
14.4
19
PDB header: transcription/dnaChain: A: PDB Molecule: protein (purine repressor);PDBTitle: purine repressor mutant-hypoxanthine-palindromic operator2 complex
30 c1bcrA_
not modelled
13.2
15
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: serine carboxypeptidase ii;PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
31 d1us7b_
not modelled
12.9
8
Fold: Hsp90 co-chaperone CDC37Superfamily: Hsp90 co-chaperone CDC37Family: Hsp90 co-chaperone CDC3732 c1us7B_
not modelled
12.9
8
PDB header: chaperoneChain: B: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: complex of hsp90 and p50
33 c1t3jA_
not modelled
12.0
18
PDB header: membrane proteinChain: A: PDB Molecule: mitofusin 1;PDBTitle: mitofusin domain hr2 v686m/i708m mutant
34 c3odnA_
not modelled
11.7
25
PDB header: membrane proteinChain: A: PDB Molecule: dally-like protein;PDBTitle: the crystal structure of drosophila dally-like protein core domain
35 c2l27A_
not modelled
11.3
50
PDB header: membrane protein, peptide binding proteiChain: A: PDB Molecule: seven transmembrane helix receptor;PDBTitle: nmr structure of the ecd1 of crf-r1 in complex with a peptide agonist
36 c3e90B_
not modelled
11.2
63
PDB header: hydrolaseChain: B: PDB Molecule: ns3 protease;PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
37 d2ijob1
not modelled
11.0
63
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases38 d2fp7b1
not modelled
10.9
63
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases39 d1xp8a2
not modelled
10.9
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain40 c3n7sB_
not modelled
10.7
27
PDB header: membrane proteinChain: B: PDB Molecule: calcitonin gene-related peptide type 1 receptor;PDBTitle: crystal structure of the ectodomain complex of the cgrp receptor, a2 class-b gpcr, reveals the site of drug antagonism
41 d2ok5a4
not modelled
10.6
50
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain42 c3q7tB_
not modelled
10.5
20
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein;PDBTitle: 2.15a resolution structure (i41 form) of the chxr receiver domain from2 chlamydia trachomatis
43 d1f37b_
not modelled
10.2
30
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioredoxin-like 2Fe-2S ferredoxin44 c3d23A_
not modelled
10.1
26
PDB header: hydrolaseChain: A: PDB Molecule: 3c-like proteinase;PDBTitle: main protease of hcov-hku1
45 c2q6fB_
not modelled
9.7
26
PDB header: hydrolaseChain: B: PDB Molecule: infectious bronchitis virus (ibv) main protease;PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
46 d1lvoa_
not modelled
9.6
26
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold47 d2vgna2
not modelled
9.4
17
Fold: Ribonuclease H-like motifSuperfamily: Translational machinery componentsFamily: ERF1/Dom34 middle domain-like48 c2re3A_
not modelled
9.3
27
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1285 family protein (spo_0140) from2 silicibacter pomeroyi dss-3 at 2.50 a resolution
49 d1q0qa1
not modelled
9.2
31
Fold: Left-handed superhelixSuperfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domainFamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain50 d1hfia_
not modelled
9.1
50
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain51 d1mo6a2
not modelled
8.9
14
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain52 d2fomb1
not modelled
8.8
38
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases53 c2d56A_
not modelled
8.6
57
PDB header: antibioticChain: A: PDB Molecule: asabf;PDBTitle: solution structure of asabf, antibacterial peptide isolated2 from a nematode, ascaris suum
54 d1dgna_
not modelled
8.6
11
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD55 d1u94a2
not modelled
8.5
17
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain56 d1p9sa_
not modelled
8.4
26
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold57 d1v86a_
not modelled
8.3
7
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related58 c1zvvA_
not modelled
8.3
29
PDB header: transcription/dnaChain: A: PDB Molecule: glucose-resistance amylase regulator;PDBTitle: crystal structure of a ccpa-crh-dna complex
59 c2auvA_
not modelled
8.3
29
PDB header: oxidoreductaseChain: A: PDB Molecule: potential nad-reducing hydrogenase subunit;PDBTitle: solution structure of hndac : a thioredoxin-like [2fe-2s]2 ferredoxin involved in the nadp-reducing hydrogenase3 complex
60 d1gx5a_
not modelled
8.2
31
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase61 d1r0ka1
not modelled
8.1
23
Fold: Left-handed superhelixSuperfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domainFamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain62 d2ffga1
not modelled
8.0
38
Fold: YkuJ-likeSuperfamily: YkuJ-likeFamily: YkuJ-like63 d1mdwa_
not modelled
7.9
21
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Calpain large subunit, catalytic domain (domain II)64 c3dl8D_
not modelled
7.8
26
PDB header: protein transportChain: D: PDB Molecule: sece;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
65 d1ubea2
not modelled
7.7
14
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain66 c2fgtA_
not modelled
7.7
29
PDB header: signaling proteinChain: A: PDB Molecule: two-component system yycf/yycg regulatoryPDBTitle: crystal structure of yych from bacillus subtilis
67 c3lkwA_
not modelled
7.4
38
PDB header: viral protein,hydrolaseChain: A: PDB Molecule: fusion protein of nonstructural protein 2b andPDBTitle: crystal structure of dengue virus 1 ns2b/ns3 protease active2 site mutant
68 d2fbea1
not modelled
7.4
22
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: SPRY domain69 c1ct9D_
not modelled
7.4
30
PDB header: ligaseChain: D: PDB Molecule: asparagine synthetase b;PDBTitle: crystal structure of asparagine synthetase b from2 escherichia coli
70 d1nb4a_
not modelled
7.3
31
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase71 d1befa_
not modelled
7.3
38
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases72 d1kgda_
not modelled
7.2
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases73 c2wl1A_
not modelled
7.1
44
PDB header: signaling proteinChain: A: PDB Molecule: pyrin;PDBTitle: pyrin pryspry domain
74 d1jgta2
not modelled
7.0
19
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases75 d1mj4a_
not modelled
6.8
44
Fold: Cytochrome b5-like heme/steroid binding domainSuperfamily: Cytochrome b5-like heme/steroid binding domainFamily: Cytochrome b576 d2iwgb1
not modelled
6.7
56
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: SPRY domain77 d1v4sa2
not modelled
6.7
28
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase78 c3rkoA_
not modelled
6.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase subunit a;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
79 d1mhqa_
not modelled
6.5
8
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain80 c3kjxD_
not modelled
6.3
19
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a transcriptional regulator, lacl2 family protein from silicibacter pomeroyi
81 d1u34a_
not modelled
6.3
63
Fold: Hormone receptor domainSuperfamily: Hormone receptor domainFamily: Hormone receptor domain82 c3ba3A_
not modelled
6.2
12
PDB header: oxidoreductaseChain: A: PDB Molecule: pyridoxamine 5'-phosphate oxidase-like protein;PDBTitle: crystal structure of pyridoxamine 5'-phosphate oxidase-like protein2 (np_783940.1) from lactobacillus plantarum at 1.55 a resolution
83 c3fotA_
not modelled
6.2
24
PDB header: transferaseChain: A: PDB Molecule: 15-o-acetyltransferase;PDBTitle: structural and functional characterization of tri3 trichothecene 15-o-2 acetyltransferase from fusarium sporotrichioides
84 c1q15A_
not modelled
6.2
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: cara;PDBTitle: carbapenam synthetase
85 c3cm8B_
not modelled
6.0
30
PDB header: rna binding protein/transferaseChain: B: PDB Molecule: peptide from rna-directed rna polymerasePDBTitle: a rna polymerase subunit structure from virus
86 c2k50A_
not modelled
6.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: replication factor a related protein;PDBTitle: solution nmr structure of the replication factor a related2 protein from methanobacterium thermoautotrophicum.3 northeast structural genomics target tr91a.
87 d1kxoa_
not modelled
6.0
8
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like88 c2bsjB_
not modelled
6.0
50
PDB header: chaperoneChain: B: PDB Molecule: chaperone protein syct;PDBTitle: native crystal structure of the type iii secretion2 chaperone syct from yersinia enterocolitica
89 d3c2wa2
not modelled
5.9
17
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: Phytochrome-specific domain90 c3kb5A_
not modelled
5.8
67
PDB header: membrane proteinChain: A: PDB Molecule: tripartite motif-containing protein 72;PDBTitle: pry-spry domain of human trim72
91 d1q15a2
not modelled
5.7
20
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases92 c3kzgB_
not modelled
5.7
8
PDB header: transport proteinChain: B: PDB Molecule: arginine 3rd transport system periplasmic bindingPDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
93 c1htrP_
not modelled
5.6
38
PDB header: aspartyl proteaseChain: P: PDB Molecule: progastricsin (pro segment);PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
94 d1scjb_
not modelled
5.6
16
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors95 c1gxsC_
not modelled
5.5
17
PDB header: lyaseChain: C: PDB Molecule: p-(s)-hydroxymandelonitrile lyase chain a;PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
96 c3jsrA_
not modelled
5.5
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: all0216 protein;PDBTitle: x-ray structure of all0216 protein from nostoc sp. pcc 7120 at the2 resolution 1.8a. northeast structural genomics consortium target3 nsr236
97 c1m1zB_
not modelled
5.5
44
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactam synthetase;PDBTitle: beta-lactam synthetase apo enzyme
98 c3fcnA_
not modelled
5.4
36
PDB header: unknown functionChain: A: PDB Molecule: an alpha-helical protein of unknown function (pfam01724);PDBTitle: crystal structure of an alpha-helical protein of unknown function2 (rru_a3208) from rhodospirillum rubrum atcc 11170 at 1.45 a3 resolution
99 d1oqja_
not modelled
5.1
14
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SAND domain