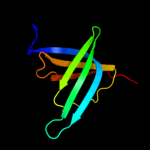
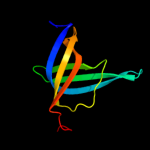
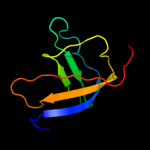
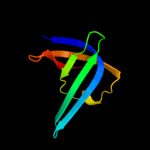
1 d1sr3a_
100.0
100
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE2 d1j6qa_
100.0
59
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE3 c1j6qA_
100.0
59
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c maturation protein e;PDBTitle: solution structure and characterization of the heme2 chaperone ccme
4 c2kctA_
99.9
25
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccme;PDBTitle: solution nmr structure of the ob-fold domain of heme2 chaperone ccme from desulfovibrio vulgaris. northeast3 structural genomics target dvr115g.
5 c3e0jG_
92.6
11
PDB header: transferaseChain: G: PDB Molecule: dna polymerase subunit delta-2;PDBTitle: x-ray structure of the complex of regulatory subunits of2 human dna polymerase delta
6 c3floG_
90.6
16
PDB header: transferaseChain: G: PDB Molecule: dna polymerase alpha subunit b;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
7 c1asyA_
86.1
11
PDB header: complex (aminoacyl-trna synthase/trna)Chain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: class ii aminoacyl transfer rna synthetases: crystal2 structure of yeast aspartyl-trna synthetase complexed with3 trna asp
8 c1eqrC_
85.0
26
PDB header: ligaseChain: C: PDB Molecule: aspartyl-trna synthetase;PDBTitle: crystal structure of free aspartyl-trna synthetase from2 escherichia coli
9 c2pi2A_
83.4
17
PDB header: replication, dna binding proteinChain: A: PDB Molecule: replication protein a 32 kda subunit;PDBTitle: full-length replication protein a subunits rpa14 and rpa32
10 d1c0aa1
83.0
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain11 c1wydB_
82.2
16
PDB header: ligaseChain: B: PDB Molecule: hypothetical aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from sulfolobus tokodaii
12 d2pi2a1
82.1
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB13 d1b8aa1
81.3
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain14 c3i7fA_
81.1
13
PDB header: ligaseChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: aspartyl trna synthetase from entamoeba histolytica
15 d1l0wa1
80.6
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain16 c3m4qA_
80.3
18
PDB header: ligaseChain: A: PDB Molecule: asparaginyl-trna synthetase, putative;PDBTitle: entamoeba histolytica asparaginyl-trna synthetase (asnrs)
17 c3kf6A_
80.1
17
PDB header: structural proteinChain: A: PDB Molecule: protein stn1;PDBTitle: crystal structure of s. pombe stn1-ten1 complex
18 c3bjuB_
72.6
18
PDB header: ligaseChain: B: PDB Molecule: lysyl-trna synthetase;PDBTitle: crystal structure of tetrameric form of human lysyl-trna2 synthetase
19 d1c9oa_
72.3
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like20 d1n9wa1
71.0
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain21 d1gm5a2
not modelled
69.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RecG "wedge" domain22 c2xgtB_
not modelled
65.1
21
PDB header: ligaseChain: B: PDB Molecule: asparaginyl-trna synthetase, cytoplasmic;PDBTitle: asparaginyl-trna synthetase from brugia malayi complexed2 with the sulphamoyl analogue of asparaginyl-adenylate
23 c3e0dA_
not modelled
63.2
30
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase iii subunit alpha;PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
24 c3e0eA_
not modelled
59.7
18
PDB header: replicationChain: A: PDB Molecule: replication protein a;PDBTitle: crystal structure of a domain of replication protein a from2 methanococcus maripaludis. northeast structural genomics3 targe mrr110b
25 c1efwA_
not modelled
57.0
21
PDB header: ligase/rnaChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from thermus2 thermophilus complexed to trnaasp from escherichia coli
26 c1b8aB_
not modelled
56.8
24
PDB header: ligaseChain: B: PDB Molecule: protein (aspartyl-trna synthetase);PDBTitle: aspartyl-trna synthetase
27 c1z9fA_
not modelled
56.7
21
PDB header: dna binding proteinChain: A: PDB Molecule: single-strand binding protein;PDBTitle: crystal structure of single stranded dna-binding protein (tm0604) from2 thermotoga maritima at 2.60 a resolution
28 d1eova1
not modelled
55.2
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain29 c1x55A_
not modelled
43.9
24
PDB header: ligaseChain: A: PDB Molecule: asparaginyl-trna synthetase;PDBTitle: crystal structure of asparaginyl-trna synthetase from pyrococcus2 horikoshii complexed with asparaginyl-adenylate analogue
30 d1bbua1
not modelled
43.7
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain31 d2es2a1
not modelled
40.4
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like32 d1o7ia_
not modelled
40.0
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB33 d2pi2e1
not modelled
40.0
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB34 c1n9wA_
not modelled
38.9
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: aspartyl-trna synthetase 2;PDBTitle: crystal structure of the non-discriminating and archaeal-2 type aspartyl-trna synthetase from thermus thermophilus
35 c3najA_
not modelled
37.8
24
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: crystal structure of a protease-resistant mutant form of human2 galectin-8
36 c3a0jB_
not modelled
37.3
23
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
37 d2hqsa2
not modelled
31.9
25
Fold: Anticodon-binding domain-likeSuperfamily: TolB, N-terminal domainFamily: TolB, N-terminal domain38 c3dm3A_
not modelled
27.5
24
PDB header: replicationChain: A: PDB Molecule: replication factor a;PDBTitle: crystal structure of a domain of a replication factor a2 protein, from methanocaldococcus jannaschii. northeast3 structural genomics target mjr118e
39 d1llca1
not modelled
25.3
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like40 c2kcmA_
not modelled
24.8
20
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: cold shock domain family protein;PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
41 c3o0rC_
not modelled
23.8
10
PDB header: immune system/oxidoreductaseChain: C: PDB Molecule: nitric oxide reductase subunit c;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
42 c2dycA_
not modelled
23.7
21
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-4;PDBTitle: crystal structure of the n-terminal domain of mouse galectin-4
43 c3b9cB_
not modelled
23.0
25
PDB header: unknown functionChain: B: PDB Molecule: hspc159;PDBTitle: crystal structure of human grp crd
44 c3nv4A_
not modelled
22.4
4
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin 9 short isoform variant;PDBTitle: crystal structure of human galectin-9 c-terminal crd in complex with2 sialyllactose
45 c1n10A_
not modelled
21.8
15
PDB header: allergenChain: A: PDB Molecule: pollen allergen phl p 1;PDBTitle: crystal structure of phl p 1, a major timothy grass pollen allergen
46 c2pqaB_
not modelled
21.8
13
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
47 c3mxnB_
not modelled
21.6
19
PDB header: replicationChain: B: PDB Molecule: recq-mediated genome instability protein 2;PDBTitle: crystal structure of the rmi core complex
48 c2yroA_
not modelled
20.7
21
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: solution structure of the c-terminal gal-bind lectin2 protein from human galectin-8
49 c1fguA_
not modelled
19.1
12
PDB header: replicationChain: A: PDB Molecule: replication protein a 70 kda dna-binding subunit;PDBTitle: ssdna-binding domain of the large subunit of replication2 protein a
50 d2gala_
not modelled
19.0
21
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)51 c2yxsA_
not modelled
18.4
29
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8 variant;PDBTitle: crystal sturcture of n-terminal domain of human galectin-8 with d-2 lactose
52 d2nn8a1
not modelled
18.2
17
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)53 c2k5nA_
not modelled
17.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cold-shock protein;PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
54 c1gm5A_
not modelled
17.5
15
PDB header: helicaseChain: A: PDB Molecule: recg;PDBTitle: structure of recg bound to three-way dna junction
55 c2yf3F_
not modelled
16.1
27
PDB header: hydrolaseChain: F: PDB Molecule: mazg-like nucleoside triphosphate pyrophosphohydrolase;PDBTitle: crystal structure of dr2231, the mazg-like protein from2 deinococcus radiodurans, complex with manganese
56 d1h95a_
not modelled
16.0
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like57 d1g6pa_
not modelled
15.8
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like58 c3e9hB_
not modelled
15.7
21
PDB header: ligaseChain: B: PDB Molecule: lysyl-trna synthetase;PDBTitle: lysyl-trna synthetase from bacillus stearothermophilus2 complexed with l-lysylsulfamoyl adenosine
59 c3kojA_
not modelled
15.5
16
PDB header: dna binding proteinChain: A: PDB Molecule: uncharacterized protein ycf41;PDBTitle: crystal structure of the ssb domain of q5n255_synp6 protein2 from synechococcus sp. northeast structural genomics3 consortium target snr59a.
60 d1lr0a_
not modelled
15.5
18
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TolA61 c2d6lX_
not modelled
14.9
25
PDB header: sugar binding proteinChain: X: PDB Molecule: lectin, galactose binding, soluble 9;PDBTitle: crystal structure of mouse galectin-9 n-terminal crd2 (crystal form 2)
62 d1n10a1
not modelled
14.2
11
Fold: C2 domain-likeSuperfamily: PHL pollen allergenFamily: PHL pollen allergen63 c2ealB_
not modelled
13.8
21
PDB header: sugar binding proteinChain: B: PDB Molecule: galectin-9;PDBTitle: crystal structure of human galectin-9 n-terminal crd in complex with2 forssman pentasaccharide
64 c2wsuB_
not modelled
13.1
16
PDB header: viral proteinChain: B: PDB Molecule: putative fiber protein;PDBTitle: galectin domain of porcine adenovirus type 4 nadc-1 isolate2 fibre
65 d1whoa_
not modelled
12.8
19
Fold: C2 domain-likeSuperfamily: PHL pollen allergenFamily: PHL pollen allergen66 d1okja1
not modelled
12.2
10
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like67 c2hczX_
not modelled
12.1
9
PDB header: allergenChain: X: PDB Molecule: beta-expansin 1a;PDBTitle: crystal structure of expb1 (zea m 1), a beta-expansin and group-12 pollen allergen from maize
68 d1l4ia2
not modelled
11.9
17
Fold: C2 domain-likeSuperfamily: Periplasmic chaperone C-domainFamily: Periplasmic chaperone C-domain69 d1ldna1
not modelled
11.2
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like70 d1jmca1
not modelled
10.9
10
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB71 d2a6aa1
not modelled
10.9
12
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like72 d1k8wa3
not modelled
10.8
10
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain73 c3lhnB_
not modelled
10.8
29
PDB header: lipid binding proteinChain: B: PDB Molecule: lipoprotein;PDBTitle: crystal structure of putative lipoprotein (np_718719.1) from2 shewanella oneidensis at 1.42 a resolution
74 c3k6oA_
not modelled
10.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein duf1344;PDBTitle: crystal structure of protein of unknown function duf13442 (yp_001299214.1) from bacteroides vulgatus atcc 8482 at 2.00 a3 resolution
75 c2qneA_
not modelled
10.6
22
PDB header: transferaseChain: A: PDB Molecule: putative methyltransferase;PDBTitle: crystal structure of putative methyltransferase (zp_00558420.1) from2 desulfitobacterium hafniense y51 at 2.30 a resolution
76 c1ynxA_
not modelled
10.5
10
PDB header: dna binding proteinChain: A: PDB Molecule: replication factor-a protein 1;PDBTitle: solution structure of dna binding domain a (dbd-a) of2 s.cerevisiae replication protein a (rpa)
77 d1krta_
not modelled
10.2
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain78 d1txya_
not modelled
9.9
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB79 d1nnxa_
not modelled
9.4
19
Fold: OB-foldSuperfamily: Hypothetical protein YgiWFamily: Hypothetical protein YgiW80 d1jnya1
not modelled
9.2
16
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors81 d1c0pa1
not modelled
9.1
15
Fold: Nucleotide-binding domainSuperfamily: Nucleotide-binding domainFamily: D-aminoacid oxidase, N-terminal domain82 c1e22A_
not modelled
9.1
24
PDB header: ligaseChain: A: PDB Molecule: lysyl-trna synthetase;PDBTitle: lysyl-trna synthetase (lysu) hexagonal form complexed with2 lysine and the non-hydrolysable atp analogue amp-pcp
83 c3eoeC_
not modelled
9.1
18
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
84 d1e1oa1
not modelled
8.8
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain85 c2jnzA_
not modelled
8.7
26
PDB header: allergenChain: A: PDB Molecule: phl p 3 allergen;PDBTitle: solution structure of phl p 3, a major allergen from2 timothy grass pollen
86 c1llcA_
not modelled
8.2
11
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: structure determination of the allosteric l-lactate dehydrogenase from2 lactobacillus casei at 3.0 angstroms resolution
87 d1mjca_
not modelled
8.1
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like88 c1x50A_
not modelled
8.0
17
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-4;PDBTitle: solution structure of the c-terminal gal-bind lectin domain2 from human galectin-4
89 c2z4hB_
not modelled
7.6
29
PDB header: signaling protein activatorChain: B: PDB Molecule: copper homeostasis protein cutf;PDBTitle: crystal structure of the cpx pathway activator nlpe from2 escherichia coli
90 c2kncA_
not modelled
7.6
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
91 d1jmca2
not modelled
7.5
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB92 d1rmda1
not modelled
6.9
13
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H293 c3e8gB_
not modelled
6.8
0
PDB header: membrane proteinChain: B: PDB Molecule: potassium channel protein;PDBTitle: crystal structure of the the open nak channel-na+/ca2+ complex
94 c2j7aC_
not modelled
6.6
19
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c quinol dehydrogenase nrfh;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
95 c2eqmA_
not modelled
6.6
14
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
96 d1o65a_
not modelled
6.5
25
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: MOSC (MOCO sulphurase C-terminal) domain97 c3e1rB_
not modelled
6.5
20
PDB header: cell cycle/transport proteinChain: B: PDB Molecule: centrosomal protein of 55 kda;PDBTitle: midbody targeting of the escrt machinery by a non-canonical2 coiled-coil in cep55
98 d1ldma1
not modelled
6.5
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like99 c1jpeA_
not modelled
6.4
21
PDB header: electron transportChain: A: PDB Molecule: dsbd-alpha;PDBTitle: crystal structure of dsbd-alpha; the n-terminal domain of2 dsbd