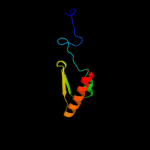
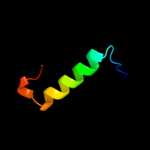
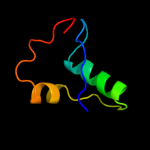
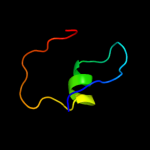
1 c2r1fB_
100.0
97
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: predicted aminodeoxychorismate lyase;PDBTitle: crystal structure of predicted aminodeoxychorismate lyase from2 escherichia coli
2 d3bz6a1
45.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PSPTO2686-like3 c2v3sB_
44.5
7
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase osr1;PDBTitle: structural insights into the recognition of substrates and2 activators by the osr1 kinase
4 d1l0wa2
38.3
29
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain5 d1gjja2
36.9
25
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain6 c3r85E_
34.5
40
PDB header: apoptosisChain: E: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
7 d1eg7a_
34.4
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like8 d1h9fa_
33.9
21
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain9 d1ztwa1
33.3
16
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Reverse transcriptase10 c3bz6A_
33.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0502 protein pspto_2686;PDBTitle: crystal structure of a conserved protein of unknown function from2 pseudomonas syringae pv. tomato str. dc3000
11 d1jeia_
32.5
29
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain12 c3r85G_
32.1
40
PDB header: apoptosisChain: G: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
13 d1e0ga_
29.6
10
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain14 d2fug13
26.9
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Nqo1 middle domain-likeFamily: Nqo1 middle domain-like15 c2exuA_
26.7
20
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation protein spt4/spt5;PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
16 c3do6B_
26.2
29
PDB header: ligaseChain: B: PDB Molecule: formate--tetrahydrofolate ligase;PDBTitle: crystal structure of putative formyltetrahydrofolate2 synthetase (tm1766) from thermotoga maritima at 1.85 a3 resolution
17 c3r85F_
26.2
41
PDB header: apoptosisChain: F: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
18 c1gjjA_
25.8
57
PDB header: membrane proteinChain: A: PDB Molecule: lap2;PDBTitle: n-terminal constant region of the nuclear envelope protein2 lap2
19 c3lk2B_
25.2
16
PDB header: protein bindingChain: B: PDB Molecule: f-actin-capping protein subunit beta isoforms 1 and 2;PDBTitle: crystal structure of capz bound to the uncapping motif from carmil
20 c3f1iH_
24.6
30
PDB header: protein bindingChain: H: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: human escrt-0 core complex
21 c2zktB_
not modelled
23.7
19
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
22 d1wxqa2
not modelled
23.1
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain23 c2dhkA_
not modelled
20.6
8
PDB header: immune systemChain: A: PDB Molecule: tbc1 domain family member 2;PDBTitle: solution structure of the ph domain of tbc1 domain family2 member 2 protein from human
24 c3r85H_
not modelled
18.8
39
PDB header: apoptosisChain: H: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
25 c3lwfD_
not modelled
18.3
15
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator (np_470886.1)2 from listeria innocua at 2.06 a resolution
26 c2yztA_
not modelled
17.8
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha1756;PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
27 d2zd1b1
not modelled
16.1
15
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Reverse transcriptase28 c3fwlA_
not modelled
15.9
11
PDB header: transferase, hydrolaseChain: A: PDB Molecule: penicillin-binding protein 1b;PDBTitle: crystal structure of the full-length transglycosylase pbp1b2 from escherichia coli
29 d1hara_
not modelled
14.2
15
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Reverse transcriptase30 d2ysca1
not modelled
13.8
100
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain31 d1nr6a_
not modelled
12.8
56
Fold: Cytochrome P450Superfamily: Cytochrome P450Family: Cytochrome P45032 c3e4eA_
not modelled
11.8
56
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450 2e1;PDBTitle: human cytochrome p450 2e1 in complex with the inhibitor 4-2 methylpyrazole
33 d1dt9a3
not modelled
10.9
9
Fold: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Superfamily: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Family: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF134 c2k9xA_
not modelled
10.7
14
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of urm1 from trypanosoma brucei
35 d1oq1a_
not modelled
10.4
41
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Hypothetical protein YesU36 c2h8kA_
not modelled
10.4
14
PDB header: transferaseChain: A: PDB Molecule: sult1c3 splice variant d;PDBTitle: human sulfotranferase sult1c3 in complex with pap
37 c2ps3A_
not modelled
9.9
7
PDB header: metal transportChain: A: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: structure and metal binding properties of znua, a2 periplasmic zinc transporter from escherichia coli
38 d1ujpa_
not modelled
9.5
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes39 c3ed4A_
not modelled
9.5
23
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
40 c1ezjA_
not modelled
9.5
12
PDB header: viral protein, transferaseChain: A: PDB Molecule: nucleocapsid phosphoprotein;PDBTitle: crystal structure of the multimerization domain of the phosphoprotein2 from sendai virus
41 d1plsa_
not modelled
9.5
21
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)42 c1d0rA_
not modelled
9.2
13
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
43 c2qieB_
not modelled
9.2
12
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase small subunit;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
44 c1us7B_
not modelled
9.2
14
PDB header: chaperoneChain: B: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: complex of hsp90 and p50
45 d1us7b_
not modelled
9.2
14
Fold: Hsp90 co-chaperone CDC37Superfamily: Hsp90 co-chaperone CDC37Family: Hsp90 co-chaperone CDC3746 d1vjka_
not modelled
8.8
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD47 c3f5fA_
not modelled
8.7
14
PDB header: transport, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein, heparanPDBTitle: crystal structure of heparan sulfate 2-o-sulfotransferase2 from gallus gallus as a maltose binding protein fusion.
48 d1rd5a_
not modelled
8.6
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes49 c3b5qB_
not modelled
8.5
18
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
50 c3s2wB_
not modelled
8.5
7
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
51 d1c0aa2
not modelled
8.4
13
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain52 d1seia_
not modelled
8.3
10
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S853 c3szzA_
not modelled
8.3
15
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
54 c3po0A_
not modelled
8.2
4
PDB header: protein bindingChain: A: PDB Molecule: small archaeal modifier protein 1;PDBTitle: crystal structure of samp1 from haloferax volcanii
55 c3ebsA_
not modelled
8.1
56
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450 2a6;PDBTitle: human cytochrome p450 2a6 i208s/i300f/g301a/s369g in complex2 with phenacetin
56 c3lggA_
not modelled
8.1
14
PDB header: hydrolaseChain: A: PDB Molecule: adenosine deaminase cecr1;PDBTitle: crystal structure of human adenosine deaminase growth factor,2 adenosine deaminase type 2 (ada2) complexed with transition state3 analogue, coformycin
57 c3jviA_
not modelled
8.1
19
PDB header: hydrolaseChain: A: PDB Molecule: protein tyrosine phosphatase;PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
58 c3bbnH_
not modelled
8.0
8
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein s8;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
59 c2ogwB_
not modelled
7.9
7
PDB header: transport proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znuaPDBTitle: structure of abc type zinc transporter from e. coli
60 d1a9xa1
not modelled
7.8
42
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain61 c3e20C_
not modelled
7.7
15
PDB header: translationChain: C: PDB Molecule: eukaryotic peptide chain release factor subunit 1;PDBTitle: crystal structure of s.pombe erf1/erf3 complex
62 c2qzuA_
not modelled
7.7
13
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
63 d2joya1
not modelled
7.5
8
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e64 c2uzhB_
not modelled
7.5
7
PDB header: lyaseChain: B: PDB Molecule: 2c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: mycobacterium smegmatis 2c-methyl-d-erythritol-2,4-2 cyclodiphosphate synthase (ispf)
65 d1gx1a_
not modelled
7.5
23
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like66 c3thaB_
not modelled
7.4
15
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
67 c2dt7A_
not modelled
7.4
67
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor 3a subunit 3;PDBTitle: solution structure of the second surp domain of human2 splicing factor sf3a120 in complex with a fragment of3 human splicing factor sf3a60
68 c2jugB_
not modelled
7.3
17
PDB header: biosynthetic proteinChain: B: PDB Molecule: tubc protein;PDBTitle: multienzyme docking in hybrid megasynthetases
69 d2ouxa2
not modelled
7.3
4
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair70 d1i94h_
not modelled
7.2
8
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S871 c2e0yB_
not modelled
7.2
19
PDB header: transferaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of the samarium derivative of mature gamma-2 glutamyltranspeptidase from escherichia coli
72 c2c1tC_
not modelled
7.0
35
PDB header: protein transport/membrane proteinChain: C: PDB Molecule: nucleoporin nup2;PDBTitle: structure of the kap60p:nup2 complex
73 c2yvsA_
not modelled
7.0
8
PDB header: oxidoreductaseChain: A: PDB Molecule: glycolate oxidase subunit glce;PDBTitle: crystal structure of glycolate oxidase subunit glce from thermus2 thermophilus hb8
74 d1s6la1
not modelled
6.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like75 c2pmpA_
not modelled
6.9
23
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from2 the isoprenoid biosynthetic pathway of arabidopsis thaliana
76 c2o1eB_
not modelled
6.8
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ycdh;PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
77 c3f0gA_
not modelled
6.7
21
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: co-crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase with cmp
78 c2nqoB_
not modelled
6.6
25
PDB header: transferaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase
79 d1iuga_
not modelled
6.6
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like80 d1ecfa1
not modelled
6.6
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)81 d1gph11
not modelled
6.5
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)82 c2iirJ_
not modelled
6.4
16
PDB header: transferaseChain: J: PDB Molecule: acetate kinase;PDBTitle: acetate kinase from a hypothermophile thermotoga maritima
83 d1an7a_
not modelled
6.4
8
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S884 c2j98A_
not modelled
6.4
100
PDB header: rna-binding proteinChain: A: PDB Molecule: replicase polyprotein 1ab;PDBTitle: human coronavirus 229e non structural protein 9 cys69ala2 mutant (nsp9)
85 c1w9qB_
not modelled
6.3
3
PDB header: cell adhesionChain: B: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the pdz tandem of human syntenin in2 complex with tnefaf peptide
86 d2nnja1
not modelled
6.3
56
Fold: Cytochrome P450Superfamily: Cytochrome P450Family: Cytochrome P45087 c3izcN_
not modelled
6.2
16
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
88 d1a6qa1
not modelled
6.0
71
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain89 d1i6ua_
not modelled
6.0
4
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S890 c3cqxD_
not modelled
5.9
33
PDB header: chaperoneChain: D: PDB Molecule: bag family molecular chaperone regulator 2;PDBTitle: chaperone complex
91 c3lp9C_
not modelled
5.9
27
PDB header: plant proteinChain: C: PDB Molecule: ls-24;PDBTitle: crystal structure of ls24, a seed albumin from lathyrus2 sativus
92 c2vqrA_
not modelled
5.8
13
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
93 c3iz5N_
not modelled
5.8
8
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
94 d2gy9h1
not modelled
5.7
16
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S895 d1b74a1
not modelled
5.7
15
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase96 d1okra_
not modelled
5.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Penicillinase repressor97 c2b5dX_
not modelled
5.6
8
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of the novel alpha-amylase amyc from thermotoga2 maritima
98 d1iv3a_
not modelled
5.6
21
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like99 d2crfa1
not modelled
5.5
8
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Ran-binding domain