1 c2vsqA_
100.0
23
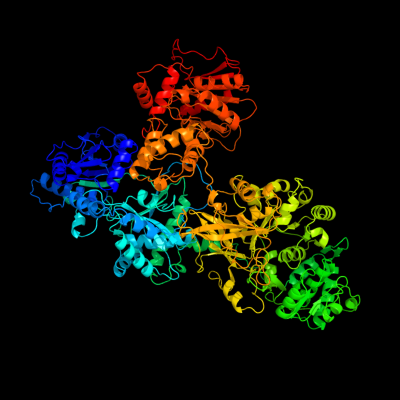
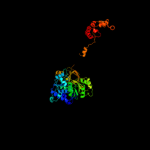
PDB header: ligaseChain: A: PDB Molecule: surfactin synthetase subunit 3;PDBTitle: structure of surfactin a synthetase c (srfa-c), a2 nonribosomal peptide synthetase termination module
2 d1pg4a_
100.0
21
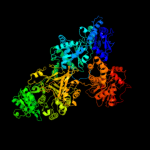
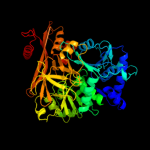
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like3 d1ry2a_
100.0
22
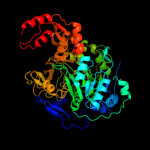
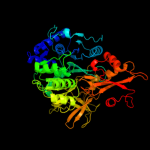
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like4 c3tsyA_
100.0
16
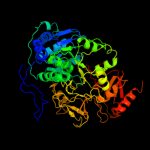
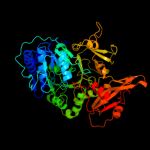
PDB header: ligase, transferaseChain: A: PDB Molecule: fusion protein 4-coumarate--coa ligase 1, resveratrolPDBTitle: 4-coumaroyl-coa ligase::stilbene synthase fusion protein
5 c3e7wA_
100.0
24
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: crystal structure of dlta: implications for the reaction2 mechanism of non-ribosomal peptide synthetase (nrps)3 adenylation domains
6 d3cw9a1
100.0
26
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like7 d1mdba_
100.0
22
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like8 c3ni2A_
100.0
20
PDB header: ligaseChain: A: PDB Molecule: 4-coumarate:coa ligase;PDBTitle: crystal structures and enzymatic mechanisms of a populus tomentosa 4-2 coumarate:coa ligase
9 c3etcB_
100.0
19
PDB header: ligaseChain: B: PDB Molecule: amp-binding protein;PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
10 d1amua_
100.0
31
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like11 c3iteB_
100.0
27
PDB header: ligaseChain: B: PDB Molecule: sidn siderophore synthetase;PDBTitle: the third adenylation domain of the fungal sidn non-2 ribosomal peptide synthetase
12 c1amuB_
100.0
30
PDB header: peptide synthetaseChain: B: PDB Molecule: gramicidin synthetase 1;PDBTitle: phenylalanine activating domain of gramicidin synthetase 12 in a complex with amp and phenylalanine
13 c3gqwB_
100.0
20
PDB header: ligaseChain: B: PDB Molecule: fatty acid amp ligase;PDBTitle: crystal structure of a fatty acid amp ligase from e. coli with an acyl2 adenylate product bound
14 c3kxwA_
100.0
17
PDB header: ligaseChain: A: PDB Molecule: saframycin mx1 synthetase b;PDBTitle: the crystal structure of fatty acid amp ligase from legionella2 pneumophila
15 c3eynB_
100.0
21
PDB header: ligaseChain: B: PDB Molecule: acyl-coenzyme a synthetase acsm2a;PDBTitle: crystal structure of human acyl-coa synthetase medium-chain2 family member 2a (l64p mutation) in a complex with coa
16 c2v7bB_
100.0
21
PDB header: ligaseChain: B: PDB Molecule: benzoate-coenzyme a ligase;PDBTitle: crystal structures of a benzoate coa ligase from2 burkholderia xenovorans lb400
17 c2d1tA_
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: luciferin 4-monooxygenase;PDBTitle: crystal structure of the thermostable japanese firefly2 luciferase red-color emission s286n mutant complexed with3 high-energy intermediate analogue
18 c3nyrA_
100.0
24
PDB header: ligaseChain: A: PDB Molecule: malonyl-coa ligase;PDBTitle: malonyl-coa ligase ternary product complex with malonyl-coa and amp2 bound
19 c3dhvA_
100.0
26
PDB header: ligaseChain: A: PDB Molecule: d-alanine-poly(phosphoribitol) ligase;PDBTitle: crystal structure of dlta protein in complex with d-alanine2 adenylate
20 d1v25a_
100.0
23
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like21 c3g7sA_
not modelled
100.0
19
PDB header: ligaseChain: A: PDB Molecule: long-chain-fatty-acid--coa ligase (fadd-1);PDBTitle: crystal structure of a long-chain-fatty-acid-coa ligase2 (fadd1) from archaeoglobus fulgidus
22 c3iplB_
not modelled
100.0
20
PDB header: ligaseChain: B: PDB Molecule: 2-succinylbenzoate--coa ligase;PDBTitle: crystal structure of o-succinylbenzoic acid-coa ligase from2 staphylococcus aureus subsp. aureus mu50
23 c3l8cA_
not modelled
100.0
24
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: structure of probable d-alanine--poly(phosphoribitol) ligase2 subunit-1 from streptococcus pyogenes
24 d1lcia_
not modelled
100.0
14
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like25 c1l5aA_
not modelled
100.0
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: amide synthase;PDBTitle: crystal structure of vibh, an nrps condensation enzyme
26 c2jgpA_
not modelled
100.0
16
PDB header: ligaseChain: A: PDB Molecule: tyrocidine synthetase 3;PDBTitle: structure of the tycc5-6 pcp-c bidomain of the tyrocidine2 synthetase tycc
27 c3o82B_
not modelled
100.0
20
PDB header: ligaseChain: B: PDB Molecule: peptide arylation enzyme;PDBTitle: structure of base n-terminal domain from acinetobacter baumannii bound2 to 5'-o-[n-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
28 c3o82A_
not modelled
100.0
21
PDB header: ligaseChain: A: PDB Molecule: peptide arylation enzyme;PDBTitle: structure of base n-terminal domain from acinetobacter baumannii bound2 to 5'-o-[n-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
29 c2xhgA_
not modelled
100.0
16
PDB header: isomeraseChain: A: PDB Molecule: tyrocidine synthetase a;PDBTitle: crystal structure of the epimerization domain from the initiation2 module of tyrocidine biosynthesis
30 c3ivrA_
not modelled
100.0
21
PDB header: ligaseChain: A: PDB Molecule: putative long-chain-fatty-acid coa ligase;PDBTitle: crystal structure of putative long-chain-fatty-acid coa ligase from2 rhodopseudomonas palustris cga009
31 c2roqA_
not modelled
100.0
100
PDB header: transferaseChain: A: PDB Molecule: enterobactin synthetase component f;PDBTitle: solution structure of the thiolation-thioesterase di-domain2 of enterobactin synthetase component f
32 c3e53A_
not modelled
100.0
21
PDB header: ligaseChain: A: PDB Molecule: fatty-acid-coa ligase fadd28;PDBTitle: crystal structure of n-terminal domain of a fatty acyl amp2 ligase faal28 from mycobacterium tuberculosis
33 c2y4oA_
not modelled
100.0
18
PDB header: ligaseChain: A: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak2 in complex with phenylacetyl adenylate
34 c2y27B_
not modelled
100.0
18
PDB header: ligaseChain: B: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak1 in complex with atp from2 burkholderia cenocepacia
35 c3qovD_
not modelled
100.0
16
PDB header: ligaseChain: D: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of a hypothetical acyl-coa ligase (bt_0428) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
36 c3fotA_
not modelled
100.0
8
PDB header: transferaseChain: A: PDB Molecule: 15-o-acetyltransferase;PDBTitle: structural and functional characterization of tri3 trichothecene 15-o-2 acetyltransferase from fusarium sporotrichioides
37 d1jmkc_
not modelled
100.0
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases38 c1q9jA_
not modelled
100.0
15
PDB header: ligaseChain: A: PDB Molecule: polyketide synthase associated protein 5;PDBTitle: structure of polyketide synthase associated protein 5 from2 mycobacterium tuberculosis
39 c2cbgA_
not modelled
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: fengycin synthetase;PDBTitle: crystal structure of the pmsf-inhibited thioesterase domain2 of the fengycin biosynthesis cluster
40 c1mo2A_
not modelled
100.0
18
PDB header: transferaseChain: A: PDB Molecule: erythronolide synthase, modules 5 and 6;PDBTitle: thioesterase domain from 6-deoxyerythronolide synthase2 (debs te), ph 8.5
41 d1mo2a_
not modelled
100.0
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases42 c3lcrA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: tautomycetin biosynthetic pks;PDBTitle: thioesterase from tautomycetin biosynthhetic pathway
43 d2h7xa1
not modelled
100.0
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases44 c2h7xA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: type i polyketide synthase pikaiv;PDBTitle: pikromycin thioesterase adduct with reduced triketide2 affinity label
45 d1l5aa1
not modelled
100.0
25
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)46 d1l5aa2
not modelled
100.0
24
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)47 c3hguB_
not modelled
100.0
18
PDB header: biosynthetic proteinChain: B: PDB Molecule: ehpf;PDBTitle: structure of phenazine antibiotic biosynthesis protein
48 d1xkta_
not modelled
100.0
21
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases49 c2px6A_
not modelled
99.9
20
PDB header: transferaseChain: A: PDB Molecule: thioesterase domain;PDBTitle: crystal structure of the thioesterase domain of human fatty2 acid synthase inhibited by orlistat
50 c3ilsA_
not modelled
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: aflatoxin biosynthesis polyketide synthase;PDBTitle: the thioesterase domain from pksa
51 c3qmwD_
not modelled
99.9
19
PDB header: hydrolaseChain: D: PDB Molecule: thioesterase;PDBTitle: redj with peg molecule bound in the active site
52 d1q9ja1
not modelled
99.9
19
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)53 c3flaB_
not modelled
99.9
16
PDB header: hydrolaseChain: B: PDB Molecule: rifr;PDBTitle: rifr - type ii thioesterase from rifamycin nrps/pks biosynthetic2 pathway - form 1
54 c2ronA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: surfactin synthetase thioesterase subunit;PDBTitle: the external thioesterase of the surfactin-synthetase
55 c2cq8A_
not modelled
99.7
11
PDB header: oxidoreductaseChain: A: PDB Molecule: 10-formyltetrahydrofolate dehydrogenase;PDBTitle: solution structure of rsgi ruh-033, a pp-binding domain of2 10-fthfdh from human cdna
56 d1q9ja2
not modelled
99.7
13
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)57 c1cr6A_
not modelled
99.7
18
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
58 c2bghA_
not modelled
99.7
14
PDB header: transferaseChain: A: PDB Molecule: vinorine synthase;PDBTitle: crystal structure of vinorine synthase
59 c2e1uA_
not modelled
99.7
12
PDB header: transferaseChain: A: PDB Molecule: acyl transferase;PDBTitle: crystal structure of dendranthema morifolium dmat
60 d2gdwa1
not modelled
99.6
29
Fold: Acyl carrier protein-likeSuperfamily: ACP-likeFamily: Peptidyl carrier domain61 c3v48B_
not modelled
99.6
13
PDB header: hydrolaseChain: B: PDB Molecule: putative aminoacrylate hydrolase rutd;PDBTitle: crystal structure of the putative alpha/beta hydrolase rutd from2 e.coli
62 c1y37A_
not modelled
99.6
16
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: structure of fluoroacetate dehalogenase from burkholderia sp. fa1
63 c3l80A_
not modelled
99.6
16
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein smu.1393c;PDBTitle: crystal structure of smu.1393c from streptococcus mutans ua159
64 c3qvmA_
not modelled
99.5
12
PDB header: hydrolaseChain: A: PDB Molecule: olei00960;PDBTitle: the structure of olei00960, a hydrolase from oleispira antarctica
65 c3om8A_
not modelled
99.5
13
PDB header: hydrolaseChain: A: PDB Molecule: probable hydrolase;PDBTitle: the crystal structure of a hydrolase from pseudomonas aeruginosa pa01
66 d2vata1
not modelled
99.5
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase67 c2qmqA_
not modelled
99.5
12
PDB header: signaling proteinChain: A: PDB Molecule: protein ndrg2;PDBTitle: crystal structure of a n-myc downstream regulated 2 protein (ndrg2,2 syld, ndr2, ai182517, au040374) from mus musculus at 1.70 a3 resolution
68 c2y6vB_
not modelled
99.5
10
PDB header: hydrolaseChain: B: PDB Molecule: peroxisomal membrane protein lpx1;PDBTitle: peroxisomal alpha-beta-hydrolase lpx1 (yor084w) from2 saccharomyces cerevisiae (crystal form i)
69 c2vavL_
not modelled
99.5
12
PDB header: transferaseChain: L: PDB Molecule: acetyl-coa--deacetylcephalosporin cPDBTitle: crystal structure of deacetylcephalosporin c2 acetyltransferase (dac-soak)
70 c3oosA_
not modelled
99.5
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase family protein;PDBTitle: the structure of an alpha/beta fold family hydrolase from bacillus2 anthracis str. sterne
71 c2xuaH_
not modelled
99.5
12
PDB header: hydrolaseChain: H: PDB Molecule: 3-oxoadipate enol-lactonase;PDBTitle: crystal structure of the enol-lactonase from burkholderia2 xenovorans lb400
72 c2r11D_
not modelled
99.5
9
PDB header: hydrolaseChain: D: PDB Molecule: carboxylesterase np;PDBTitle: crystal structure of putative hydrolase (2632844) from2 bacillus subtilis at 1.96 a resolution
73 c3kdaB_
not modelled
99.5
12
PDB header: hydrolaseChain: B: PDB Molecule: cftr inhibitory factor (cif);PDBTitle: crystal structure of the cftr inhibitory factor cif with the h269a2 mutation
74 c3ibtA_
not modelled
99.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase;PDBTitle: structure of 1h-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (qdo)
75 c3e0xB_
not modelled
99.4
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lipase-esterase related protein;PDBTitle: the crystal structure of a lipase-esterase related protein2 from clostridium acetobutylicum atcc 824
76 c2wj4B_
not modelled
99.4
11
PDB header: oxidoreductaseChain: B: PDB Molecule: 1h-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase;PDBTitle: crystal structure of the cofactor-devoid 1-h-3-hydroxy-4-2 oxoquinaldine 2,4-dioxygenase (hod) from arthrobacter3 nitroguajacolicus ru61a anaerobically complexed with its4 natural substrate 1-h-3-hydroxy-4-oxoquinaldine
77 c2fq1A_
not modelled
99.4
22
PDB header: hydrolaseChain: A: PDB Molecule: isochorismatase;PDBTitle: crystal structure of the two-domain non-ribosomal peptide synthetase2 entb containing isochorismate lyase and aryl-carrier protein domains
78 c3i1iA_
not modelled
99.4
16
PDB header: transferaseChain: A: PDB Molecule: homoserine o-acetyltransferase;PDBTitle: x-ray crystal structure of homoserine o-acetyltransferase from2 bacillus anthracis
79 c3fsgC_
not modelled
99.4
17
PDB header: hydrolaseChain: C: PDB Molecule: alpha/beta superfamily hydrolase;PDBTitle: crystal structure of alpha/beta superfamily hydrolase from oenococcus2 oeni psu-1
80 d1mj5a_
not modelled
99.4
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase81 d2b61a1
not modelled
99.4
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase82 c3bf7B_
not modelled
99.4
13
PDB header: hydrolaseChain: B: PDB Molecule: esterase ybff;PDBTitle: 1.1 resolution structure of ybff, a new esterase from2 escherichia coli: a unique substrate-binding crevice3 generated by domain arrangement
83 d1m33a_
not modelled
99.4
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Biotin biosynthesis protein BioH84 c2cjpA_
not modelled
99.4
12
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: structure of potato (solanum tuberosum) epoxide hydrolase i2 (steh1)
85 c2ju2A_
not modelled
99.4
25
PDB header: transferaseChain: A: PDB Molecule: erythronolide synthase;PDBTitle: minimized mean solution structure of the acyl carrier2 protein domain from module 2 of 6-deoxyerythronolide b3 synthase (debs)
86 d1ehya_
not modelled
99.4
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase87 c2liuA_
not modelled
99.3
11
PDB header: transferaseChain: A: PDB Molecule: cura;PDBTitle: nmr structure of holo-acpi domain from cura module from lyngbya2 majuscula
88 d1a8qa_
not modelled
99.3
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase89 d1e89a_
not modelled
99.3
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like90 d1hlga_
not modelled
99.3
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase91 d1hkha_
not modelled
99.3
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase92 d2pl5a1
not modelled
99.3
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase93 d1cr6a2
not modelled
99.3
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase94 d1mtza_
not modelled
99.3
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Proline iminopeptidase-like95 d1zd3a2
not modelled
99.3
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase96 c2pseA_
not modelled
99.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: renilla-luciferin 2-monooxygenase;PDBTitle: crystal structures of the luciferase and green fluorescent2 protein from renilla reniformis
97 c3kxpD_
not modelled
99.2
15
PDB header: hydrolaseChain: D: PDB Molecule: alpha-(n-acetylaminomethylene)succinic acidPDBTitle: crystal structure of e-2-(acetamidomethylene)succinate2 hydrolase
98 d1k8qa_
not modelled
99.2
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase99 c3bwxA_
not modelled
99.2
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase;PDBTitle: crystal structure of an alpha/beta hydrolase (yp_496220.1) from2 novosphingobium aromaticivorans dsm 12444 at 1.50 a resolution
100 c2xr7A_
not modelled
99.2
10
PDB header: transferaseChain: A: PDB Molecule: malonyltransferase;PDBTitle: crystal structure of nicotiana tabacum malonyltransferase (ntmat1)2 complexed with malonyl-coa
101 c2xmzA_
not modelled
99.2
14
PDB header: lyaseChain: A: PDB Molecule: hydrolase, alpha/beta hydrolase fold family;PDBTitle: structure of menh from s. aureus
102 d1brta_
not modelled
99.2
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase103 c2afdA_
not modelled
99.2
20
PDB header: ligand binding proteinChain: A: PDB Molecule: protein asl1650;PDBTitle: solution structure of asl1650, an acyl carrier protein from anabaena2 sp. pcc 7120 with a variant phosphopantetheinylation-site sequence
104 c3r3xA_
not modelled
99.2
12
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: crystal structure of the fluoroacetate dehalogenase rpa1163 -2 asp110asn/bromoacetate
105 c3laxA_
not modelled
99.2
16
PDB header: ligaseChain: A: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: the crystal structure of a domain of phenylacetate-coenzyme2 a ligase from bacteroides vulgatus atcc 8482
106 c1u2eA_
not modelled
99.2
16
PDB header: hydrolaseChain: A: PDB Molecule: 2-hydroxy-6-ketonona-2,4-dienedioic acidPDBTitle: crystal structure of the c-c bond hydrolase mhpc
107 d3c70a1
not modelled
99.2
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like108 c3r0vA_
not modelled
99.2
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase fold protein;PDBTitle: the crystal structure of an alpha/beta hydrolase from sphaerobacter2 thermophilus dsm 20745.
109 d1a8sa_
not modelled
99.2
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase110 d1xkla_
not modelled
99.2
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like111 d2rhwa1
not modelled
99.2
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase112 c3fobA_
not modelled
99.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: bromoperoxidase;PDBTitle: crystal structure of bromoperoxidase from bacillus anthracis
113 d1c4xa_
not modelled
99.1
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carbon-carbon bond hydrolase114 d1va4a_
not modelled
99.1
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase115 c3e3aA_
not modelled
99.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: possible peroxidase bpoc;PDBTitle: the structure of rv0554 from mycobacterium tuberculosis
116 c3u1tA_
not modelled
99.1
16
PDB header: hydrolaseChain: A: PDB Molecule: dmma haloalkane dehalogenase;PDBTitle: haloalkane dehalogenase, dmma, of marine microbial origin
117 c3qyjB_
not modelled
99.1
14
PDB header: hydrolaseChain: B: PDB Molecule: alr0039 protein;PDBTitle: crystal structure of alr0039, a putative alpha/beta hydrolase from2 nostoc sp pcc 7120.
118 d1qo7a_
not modelled
99.1
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase119 d1ispa_
not modelled
99.1
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase120 c2qvbA_
not modelled
99.1
15
PDB header: hydrolaseChain: A: PDB Molecule: haloalkane dehalogenase 3;PDBTitle: crystal structure of haloalkane dehalogenase rv2579 from mycobacterium2 tuberculosis