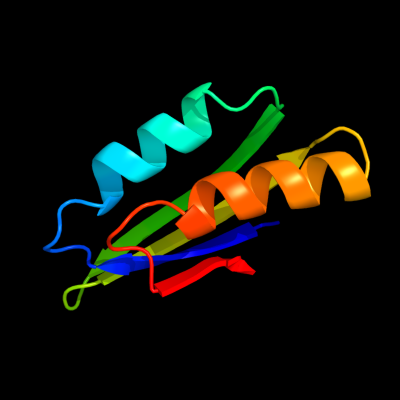
1 c3ibwA_
97.1
16
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
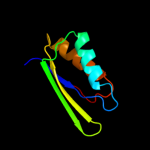
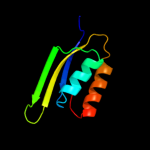
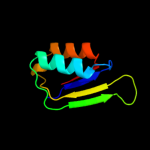
2 d1sc6a3
95.6
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain3 d1u8sa2
95.5
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor4 d1ygya3
94.3
10
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain5 d2f1fa1
94.0
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like6 d2fgca2
93.3
12
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like7 d2pc6a2
92.5
12
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like8 c2f1fA_
92.4
16
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase isozyme iii small subunit;PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
9 c2fgcA_
91.2
12
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase, small subunit;PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
10 c1ygyA_
90.0
10
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
11 c2pc6C_
89.6
12
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
12 c1rwuA_
88.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0250 protein ybed;PDBTitle: solution structure of conserved protein ybed from e. coli
13 d1rwua_
88.5
16
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: YbeD-like14 d2joqa1
80.6
12
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: HP0495-like15 d1kona_
74.9
13
Fold: YebC-likeSuperfamily: YebC-likeFamily: YebC-like16 c2e1cA_
74.3
10
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
17 c1y7pB_
73.7
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein af1403;PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
18 d2cyya2
68.6
12
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain19 c2phmA_
65.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (phenylalanine-4-hydroxylase);PDBTitle: structure of phenylalanine hydroxylase dephosphorylated
20 c2jsxA_
63.5
9
PDB header: chaperoneChain: A: PDB Molecule: protein napd;PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
21 d1lfpa_
not modelled
62.1
13
Fold: YebC-likeSuperfamily: YebC-likeFamily: YebC-like22 c2ew9A_
not modelled
56.3
8
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of apowln5-6
23 d1zpva1
not modelled
55.9
5
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like24 d1mw7a_
not modelled
55.7
16
Fold: YebC-likeSuperfamily: YebC-likeFamily: YebC-like25 c3k5pA_
not modelled
54.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
26 c2djwF_
not modelled
54.1
9
PDB header: unknown functionChain: F: PDB Molecule: probable transcriptional regulator, asnc family;PDBTitle: crystal structure of ttha0845 from thermus thermophilus hb8
27 c2e1aD_
not modelled
52.1
6
PDB header: transcriptionChain: D: PDB Molecule: 75aa long hypothetical regulatory protein asnc;PDBTitle: crystal structure of ffrp-dm1
28 d1qupa2
not modelled
52.0
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain29 d1lk5a2
not modelled
50.5
14
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain30 d1i1ga2
not modelled
49.9
9
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain31 c1u8sB_
not modelled
49.7
17
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
32 c2zbcH_
not modelled
47.6
9
PDB header: transcriptionChain: H: PDB Molecule: 83aa long hypothetical transcriptional regulator asnc;PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
33 d1u8sa1
not modelled
46.6
5
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor34 c1i1gA_
not modelled
40.7
9
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
35 c2qmxB_
not modelled
39.2
9
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
36 d3bpda1
not modelled
38.8
16
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like37 d2f06a2
not modelled
38.5
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like38 c1ybaC_
not modelled
38.2
19
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: the active form of phosphoglycerate dehydrogenase
39 d1tdja2
not modelled
36.5
10
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Allosteric threonine deaminase C-terminal domain40 c2x3dC_
not modelled
36.3
35
PDB header: unknown functionChain: C: PDB Molecule: sso6206;PDBTitle: crystal structure of sso6206 from sulfolobus solfataricus p2
41 d1phza1
not modelled
34.9
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain42 d2raqa1
not modelled
34.0
24
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like43 c2crlA_
not modelled
33.2
18
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
44 c3mtjA_
not modelled
32.0
8
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
45 c3mwbA_
not modelled
31.5
12
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
46 d1m0sa2
not modelled
30.5
13
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain47 d1o8ba2
not modelled
28.1
10
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain48 d2cg4a2
not modelled
27.8
21
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain49 d1p6ta1
not modelled
26.3
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain50 c2e7xA_
not modelled
26.3
7
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
51 c3hyjD_
not modelled
26.1
12
PDB header: transcription regulatorChain: D: PDB Molecule: protein duf199/whia;PDBTitle: crystal structure of the n-terminal laglidadg domain of duf199/whia
52 c2gqqB_
not modelled
26.1
11
PDB header: transcriptionChain: B: PDB Molecule: leucine-responsive regulatory protein;PDBTitle: crystal structure of e. coli leucine-responsive regulatory protein2 (lrp)
53 c3n0vD_
not modelled
25.4
5
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
54 c3dxsX_
not modelled
25.2
12
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
55 d2nzca1
not modelled
25.2
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: TM1266-like56 c2l3mA_
not modelled
25.0
14
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
57 d2qmwa2
not modelled
24.8
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain58 c2aj1A_
not modelled
24.5
20
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
59 c3luyA_
not modelled
24.3
14
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
60 c2ldiA_
not modelled
24.2
15
PDB header: hydrolaseChain: A: PDB Molecule: zinc-transporting atpase;PDBTitle: nmr solution structure of ziaan sub mutant
61 d2qifa1
not modelled
23.8
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain62 d1s6ua_
not modelled
23.5
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain63 c2p6tH_
not modelled
23.3
8
PDB header: transcriptionChain: H: PDB Molecule: transcriptional regulator, lrp/asnc family;PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
64 c2bj3D_
not modelled
22.5
14
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
65 c2yy3B_
not modelled
22.1
10
PDB header: translationChain: B: PDB Molecule: elongation factor 1-beta;PDBTitle: crystal structure of translation elongation factor ef-1 beta from2 pyrococcus horikoshii
66 d1q8la_
not modelled
21.4
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain67 d1uj4a2
not modelled
21.2
15
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain68 d1cc8a_
not modelled
20.9
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain69 d1p6ta2
not modelled
20.5
18
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain70 c2cg4B_
not modelled
18.8
9
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
71 c2ia0A_
not modelled
17.3
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
72 c2rogA_
not modelled
17.0
19
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
73 d1sb6a_
not modelled
16.8
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain74 c1yjrA_
not modelled
16.5
11
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
75 d1osda_
not modelled
16.1
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain76 c3i4pA_
not modelled
16.0
4
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, asnc family;PDBTitle: crystal structure of asnc family transcriptional regulator from2 agrobacterium tumefaciens
77 c2gcfA_
not modelled
15.8
9
PDB header: hydrolaseChain: A: PDB Molecule: cation-transporting atpase pacs;PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
78 d1u0sa_
not modelled
15.7
16
Fold: Ferredoxin-likeSuperfamily: CheY-binding domain of CheAFamily: CheY-binding domain of CheA79 c2vbzA_
not modelled
15.3
15
PDB header: dna-binding proteinChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
80 d1afia_
not modelled
14.2
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain81 c1lk5C_
not modelled
14.0
13
PDB header: isomeraseChain: C: PDB Molecule: d-ribose-5-phosphate isomerase;PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
82 d2ggpb1
not modelled
13.6
18
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain83 c1vi7A_
not modelled
13.3
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yigz;PDBTitle: crystal structure of an hypothetical protein
84 d2f1fa2
not modelled
13.1
6
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like85 c2ca9B_
not modelled
12.9
16
PDB header: transcriptional regulationChain: B: PDB Molecule: putative nickel-responsive regulator;PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
86 d2i5nl1
not modelled
12.7
21
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits87 c2ofhX_
not modelled
12.5
15
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
88 d2fmra_
not modelled
12.4
13
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)89 c2k2pA_
not modelled
12.0
5
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
90 c2kt2A_
not modelled
11.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
91 c2y3yC_
not modelled
11.2
16
PDB header: transcriptionChain: C: PDB Molecule: putative nickel-responsive regulator;PDBTitle: holo-ni(ii) hpnikr is a symmetric tetramer containing four2 canonic square-planar ni(ii) ions at physiological ph
92 c1yg0A_
not modelled
11.2
16
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
93 d1q5ya_
not modelled
11.1
6
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Nickel responsive regulator NikR, C-terminal domain94 d2pc6a1
not modelled
11.0
11
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like95 c2cveA_
not modelled
10.9
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1053;PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
96 c2w2mP_
not modelled
10.6
29
PDB header: hydrolase/receptorChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: wt pcsk9-deltac bound to wt egf-a of ldlr
97 c2pmwA_
not modelled
10.6
29
PDB header: hydrolaseChain: A: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: the crystal structure of proprotein convertase subtilisin2 kexin type 9 (pcsk9)
98 c2p4eP_
not modelled
10.6
29
PDB header: hydrolaseChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: crystal structure of pcsk9
99 d2cu6a1
not modelled
10.3
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like