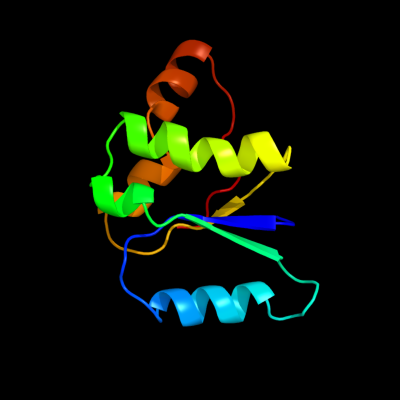
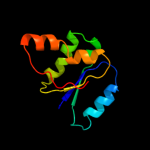
| 1 | c3kf6A_
|
|
 |
96.8 |
17 |
PDB header:structural protein
Chain: A: PDB Molecule:protein stn1;
PDBTitle: crystal structure of s. pombe stn1-ten1 complex
|
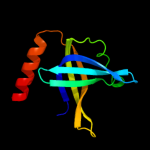
| 2 | c1z9fA_
|
|
 |
96.4 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-strand binding protein;
PDBTitle: crystal structure of single stranded dna-binding protein (tm0604) from2 thermotoga maritima at 2.60 a resolution
|
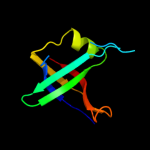
| 3 | c3bfjK_
|
|
 |
96.3 |
25 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:1,3-propanediol oxidoreductase;
PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
|
| 4 | c3f2cA_
|
|
 |
95.9 |
20 |
PDB header:transferase/dna
Chain: A: PDB Molecule:geobacillus kaustophilus dna polc;
PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
|
| 5 | c3ox4D_
|
|
 |
95.9 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 2;
PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
|
| 6 | d1oj7a_
|
|
 |
95.9 |
18 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 7 | d1vlja_
|
|
 |
95.9 |
19 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 8 | d1v1qa_
|
|
 |
95.7 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 9 | c2iheA_
|
|
 |
95.4 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: crystal structure of wild-type single-stranded dna binding protein2 from thermus aquaticus
|
| 10 | c3okfA_
|
|
 |
95.4 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
|
| 11 | d1rrma_
|
|
 |
95.4 |
21 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 12 | d1o2da_
|
|
 |
95.3 |
26 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 13 | d1bbua1
|
|
 |
95.2 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 14 | c3qvjB_
|
|
 |
95.2 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 15 | c3e0eA_
|
|
 |
95.1 |
24 |
PDB header:replication
Chain: A: PDB Molecule:replication protein a;
PDBTitle: crystal structure of a domain of replication protein a from2 methanococcus maripaludis. northeast structural genomics3 targe mrr110b
|
| 16 | d1l0wa1
|
|
 |
95.0 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 17 | d1gm5a2
|
|
 |
95.0 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RecG "wedge" domain |
| 18 | c3bjuB_
|
|
 |
95.0 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:lysyl-trna synthetase;
PDBTitle: crystal structure of tetrameric form of human lysyl-trna2 synthetase
|
| 19 | c3hl0B_
|
|
 |
94.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:maleylacetate reductase;
PDBTitle: crystal structure of maleylacetate reductase from agrobacterium2 tumefaciens
|
| 20 | d1e1oa1
|
|
 |
94.7 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 21 | c3jzdA_ |
|
not modelled |
94.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-containing alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehedrogenase (yp_298327.1) from2 ralstonia eutropha jmp134 at 2.10 a resolution
|
| 22 | d1jq5a_ |
|
not modelled |
94.7 |
22 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 23 | d1n9wa1 |
|
not modelled |
94.6 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 24 | c3tqyA_ |
|
not modelled |
94.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: structure of a single-stranded dna-binding protein (ssb), from2 coxiella burnetii
|
| 25 | d1c0aa1 |
|
not modelled |
94.5 |
12 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 26 | c3eivB_ |
|
not modelled |
94.4 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:single-stranded dna-binding protein 2;
PDBTitle: crystal structure of single-stranded dna-binding protein2 from streptomyces coelicolor
|
| 27 | c3gjzB_ |
|
not modelled |
94.4 |
8 |
PDB header:immune system
Chain: B: PDB Molecule:microcin immunity protein mccf;
PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
|
| 28 | d1ujna_ |
|
not modelled |
94.3 |
13 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Dehydroquinate synthase, DHQS |
| 29 | c2ihfA_ |
|
not modelled |
94.1 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: crystal structure of deletion mutant delta 228-252 r190a of the2 single-stranded dna binding protein from thermus aquaticus
|
| 30 | c3fduF_ |
|
not modelled |
93.7 |
17 |
PDB header:isomerase
Chain: F: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase/isomerase from2 acinetobacter baumannii
|
| 31 | c2xgtB_ |
|
not modelled |
93.6 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:asparaginyl-trna synthetase, cytoplasmic;
PDBTitle: asparaginyl-trna synthetase from brugia malayi complexed2 with the sulphamoyl analogue of asparaginyl-adenylate
|
| 32 | c3k8aA_ |
|
not modelled |
93.6 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative primosomal replication protein;
PDBTitle: neisseria gonorrhoeae prib
|
| 33 | d1eova1 |
|
not modelled |
93.4 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 34 | d1ue1a_ |
|
not modelled |
93.4 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 35 | c2eq5D_ |
|
not modelled |
93.4 |
22 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
| 36 | c3en2A_ |
|
not modelled |
93.4 |
5 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable primosomal replication protein n;
PDBTitle: three-dimensional structure of the protein prib from2 ralstonia solanacearum at the resolution 2.3a. northeast3 structural genomics consortium target rsr213c.
|
| 37 | d2nu7b1
|
|
 |
93.4 |
18 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 38 | c2vlbC_ |
|
not modelled |
93.3 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 39 | d1txya_ |
|
not modelled |
93.2 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 40 | c3moyA_ |
|
not modelled |
93.2 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:probable enoyl-coa hydratase;
PDBTitle: crystal structure of probable enoyl-coa hydratase from mycobacterium2 smegmatis
|
| 41 | c2pi2A_ |
|
not modelled |
93.0 |
19 |
PDB header:replication, dna binding protein
Chain: A: PDB Molecule:replication protein a 32 kda subunit;
PDBTitle: full-length replication protein a subunits rpa14 and rpa32
|
| 42 | c3gkbA_ |
|
not modelled |
92.8 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase from streptomyces2 avermitilis
|
| 43 | c2vw9B_ |
|
not modelled |
92.6 |
21 |
PDB header:dna-binding protein
Chain: B: PDB Molecule:single-stranded dna binding protein;
PDBTitle: single stranded dna binding protein complex from2 helicobacter pylori
|
| 44 | c2qq3F_ |
|
not modelled |
92.6 |
20 |
PDB header:lyase
Chain: F: PDB Molecule:enoyl-coa hydratase subunit i;
PDBTitle: crystal structure of enoyl-coa hydrates subunit i (gk_2039)2 other form from geobacillus kaustophilus hta426
|
| 45 | c3uhjE_ |
|
not modelled |
92.6 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:probable glycerol dehydrogenase;
PDBTitle: crystal structure of a probable glycerol dehydrogenase from2 sinorhizobium meliloti 1021
|
| 46 | d2pi2a1 |
|
not modelled |
92.5 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 47 | c3ce9A_ |
|
not modelled |
92.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase (np_348253.1) from2 clostridium acetobutylicum at 2.37 a resolution
|
| 48 | c3iv7B_ |
|
not modelled |
92.1 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase iv;
PDBTitle: crystal structure of iron-containing alcohol dehydrogenase2 (np_602249.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.07 a resolution
|
| 49 | c1ta9A_ |
|
not modelled |
92.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
|
| 50 | c3e9hB_ |
|
not modelled |
91.8 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:lysyl-trna synthetase;
PDBTitle: lysyl-trna synthetase from bacillus stearothermophilus2 complexed with l-lysylsulfamoyl adenosine
|
| 51 | d1se8a_ |
|
not modelled |
91.8 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 52 | c1se8A_ |
|
not modelled |
91.8 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-strand binding protein;
PDBTitle: structure of single-stranded dna-binding protein (ssb) from d.2 radiodurans
|
| 53 | c3mybA_ |
|
not modelled |
91.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase mycobacterium smegmatis
|
| 54 | c1gm5A_ |
|
not modelled |
91.2 |
18 |
PDB header:helicase
Chain: A: PDB Molecule:recg;
PDBTitle: structure of recg bound to three-way dna junction
|
| 55 | d1q52a_ |
|
not modelled |
91.2 |
23 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 56 | c2ej5B_ |
|
not modelled |
91.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase subunit ii;
PDBTitle: crystal structure of gk2038 protein (enoyl-coa hydratase subunit ii)2 from geobacillus kaustophilus
|
| 57 | c2iexA_ |
|
not modelled |
91.0 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxynapthoic acid synthetase;
PDBTitle: crystal structure of dihydroxynapthoic acid synthetase (gk2873) from2 geobacillus kaustophilus hta426
|
| 58 | d1b8aa1 |
|
not modelled |
90.9 |
12 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 59 | c3clhA_ |
|
not modelled |
90.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of 3-dehydroquinate synthase (dhqs)from2 helicobacter pylori
|
| 60 | c1rjnC_ |
|
not modelled |
90.6 |
21 |
PDB header:lyase
Chain: C: PDB Molecule:menb;
PDBTitle: the crystal structure of menb (rv0548c) from mycobacterium2 tuberculosis in complex with the coa portion of naphthoyl coa
|
| 61 | d1uiya_ |
|
not modelled |
90.5 |
23 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 62 | c2x58B_ |
|
not modelled |
90.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peroxisomal bifunctional enzyme;
PDBTitle: the crystal structure of mfe1 liganded with coa
|
| 63 | d1ef8a_ |
|
not modelled |
90.4 |
22 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 64 | d1qvca_ |
|
not modelled |
90.3 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 65 | c3dm3A_ |
|
not modelled |
90.3 |
23 |
PDB header:replication
Chain: A: PDB Molecule:replication factor a;
PDBTitle: crystal structure of a domain of a replication factor a2 protein, from methanocaldococcus jannaschii. northeast3 structural genomics target mjr118e
|
| 66 | d1wdka4 |
|
not modelled |
90.3 |
21 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 67 | c2issF_ |
|
not modelled |
90.2 |
24 |
PDB header:lyase, transferase
Chain: F: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
|
| 68 | c3h0uB_ |
|
not modelled |
90.2 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase from2 streptomyces avermitilis
|
| 69 | d2nv0a1 |
|
not modelled |
90.2 |
26 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 70 | c2ppyE_ |
|
not modelled |
90.0 |
15 |
PDB header:lyase
Chain: E: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydrates (gk_1992) from geobacillus2 kaustophilus hta426
|
| 71 | c1ue7A_ |
|
not modelled |
90.0 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-strand binding protein;
PDBTitle: crystal structure of the single-stranded dna-binding2 protein from mycobacterium tuberculosis
|
| 72 | d1eyga_ |
|
not modelled |
89.9 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 73 | d1szoa_ |
|
not modelled |
89.9 |
31 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 74 | c2vx2D_ |
|
not modelled |
89.8 |
26 |
PDB header:isomerase
Chain: D: PDB Molecule:enoyl-coa hydratase domain-containing protein 3;
PDBTitle: crystal structure of human enoyl coenzyme a hydratase2 domain-containing protein 3 (echdc3)
|
| 75 | c3h81A_ |
|
not modelled |
89.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase echa8;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium2 tuberculosis
|
| 76 | d3ulla_ |
|
not modelled |
89.8 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 77 | d2auna2 |
|
not modelled |
89.7 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 78 | d2a7ka1 |
|
not modelled |
89.7 |
22 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 79 | c2q35A_ |
|
not modelled |
89.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:curf;
PDBTitle: crystal structure of the y82f variant of ech2 decarboxylase domain of2 curf from lyngbya majuscula
|
| 80 | c3rf7A_ |
|
not modelled |
89.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-containing alcohol dehydrogenase;
PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
|
| 81 | c2gruB_ |
|
not modelled |
89.5 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:2-deoxy-scyllo-inosose synthase;
PDBTitle: crystal structure of 2-deoxy-scyllo-inosose synthase2 complexed with carbaglucose-6-phosphate, nad+ and co2+
|
| 82 | c2fbmB_ |
|
not modelled |
89.5 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:y chromosome chromodomain protein 1, telomeric isoform b;
PDBTitle: acetyltransferase domain of cdy1
|
| 83 | c3lkeA_ |
|
not modelled |
89.4 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from bacillus2 halodurans
|
| 84 | c3hrxD_ |
|
not modelled |
89.3 |
24 |
PDB header:lyase
Chain: D: PDB Molecule:probable enoyl-coa hydratase;
PDBTitle: crystal structure of phenylacetic acid degradation protein paag
|
| 85 | d1jvna2 |
|
not modelled |
89.1 |
26 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 86 | c3g64A_ |
|
not modelled |
89.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of putative enoyl-coa hydratase from streptomyces2 coelicolor a3(2)
|
| 87 | c3omeE_ |
|
not modelled |
89.0 |
26 |
PDB header:lyase
Chain: E: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase from mycobacterium2 smegmatis
|
| 88 | c3rsiA_ |
|
not modelled |
88.9 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: the structure of a putative enoyl-coa hydratase/isomerase from2 mycobacterium abscessus atcc 19977 / dsm 44196
|
| 89 | d1hzda_ |
|
not modelled |
88.8 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 90 | d1zl0a2 |
|
not modelled |
88.5 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 91 | c3pgzB_ |
|
not modelled |
88.4 |
12 |
PDB header:dna binding protein
Chain: B: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: crystal structure of a single strand binding protein (ssb) from2 bartonella henselae
|
| 92 | c2k50A_ |
|
not modelled |
88.4 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:replication factor a related protein;
PDBTitle: solution nmr structure of the replication factor a related2 protein from methanobacterium thermoautotrophicum.3 northeast structural genomics target tr91a.
|
| 93 | c2kenA_ |
|
not modelled |
88.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein;
PDBTitle: solution nmr structure of the ob domain (residues 67-166)2 of mm0293 from methanosarcina mazei. northeast structural3 genomics consortium target mar214a.
|
| 94 | c3kojA_ |
|
not modelled |
88.0 |
5 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uncharacterized protein ycf41;
PDBTitle: crystal structure of the ssb domain of q5n255_synp6 protein2 from synechococcus sp. northeast structural genomics3 consortium target snr59a.
|
| 95 | c2j5gL_ |
|
not modelled |
87.9 |
29 |
PDB header:hydrolase
Chain: L: PDB Molecule:alr4455 protein;
PDBTitle: the native structure of a beta-diketone hydrolase from the2 cyanobacterium anabaena sp. pcc 7120
|
| 96 | d1xx4a_ |
|
not modelled |
87.9 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 97 | c3kklA_ |
|
not modelled |
87.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable chaperone protein hsp33;
PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
|
| 98 | c1zrsB_ |
|
not modelled |
87.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: wild-type ld-carboxypeptidase
|
| 99 | c3bptA_ |
|
not modelled |
87.8 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-hydroxyisobutyryl-coa hydrolase;
PDBTitle: crystal structure of human beta-hydroxyisobutyryl-coa hydrolase in2 complex with quercetin
|
| 100 | c3lgjA_ |
|
not modelled |
87.6 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: crystal structure of single-stranded binding protein (ssb) from2 bartonella henselae
|
| 101 | d1rjma_ |
|
not modelled |
87.4 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 102 | d1nzya_ |
|
not modelled |
87.3 |
17 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 103 | c3kqfC_ |
|
not modelled |
86.8 |
17 |
PDB header:isomerase
Chain: C: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: 1.8 angstrom resolution crystal structure of enoyl-coa hydratase from2 bacillus anthracis.
|
| 104 | d1eucb1 |
|
not modelled |
86.8 |
12 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 105 | c3njbA_ |
|
not modelled |
86.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium smegmatis,2 iodide soak
|
| 106 | d1t3ta2 |
|
not modelled |
86.8 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 107 | c3peaD_ |
|
not modelled |
86.6 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of enoyl-coa hydratase from bacillus anthracis str.2 'ames ancestor'
|
| 108 | c3p5mB_ |
|
not modelled |
86.6 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of an enoyl-coa hydratase/isomerase from2 mycobacterium avium
|
| 109 | d1krta_ |
|
not modelled |
86.6 |
12 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 110 | d2fw2a1 |
|
not modelled |
86.6 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 111 | c1n9wA_ |
|
not modelled |
86.2 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:aspartyl-trna synthetase 2;
PDBTitle: crystal structure of the non-discriminating and archaeal-2 type aspartyl-trna synthetase from thermus thermophilus
|
| 112 | c2ywjA_ |
|
not modelled |
86.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: crystal structure of uncharacterized conserved protein from2 methanocaldococcus jannaschii
|
| 113 | c3q1tB_ |
|
not modelled |
86.2 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium avium
|
| 114 | c3l3sF_ |
|
not modelled |
86.2 |
19 |
PDB header:isomerase
Chain: F: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of an enoyl-coa hydrotase/isomerase family2 protein from silicibacter pomeroyi
|
| 115 | c3sllC_ |
|
not modelled |
86.1 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:probable enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 116 | c3m4qA_ |
|
not modelled |
85.9 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:asparaginyl-trna synthetase, putative;
PDBTitle: entamoeba histolytica asparaginyl-trna synthetase (asnrs)
|
| 117 | c2d3tB_ |
|
not modelled |
85.8 |
20 |
PDB header:lyase, oxidoreductase/transferase
Chain: B: PDB Molecule:fatty oxidation complex alpha subunit;
PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
|
| 118 | d2f6qa1 |
|
not modelled |
85.8 |
17 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 119 | c1e22A_ |
|
not modelled |
85.7 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:lysyl-trna synthetase;
PDBTitle: lysyl-trna synthetase (lysu) hexagonal form complexed with2 lysine and the non-hydrolysable atp analogue amp-pcp
|
| 120 | c3r6hA_ |
|
not modelled |
85.5 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase, echa3;
PDBTitle: crystal structure of an enoyl-coa hydratase (echa3) from mycobacterium2 marinum
|