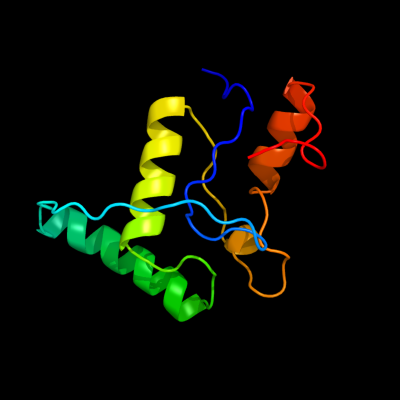
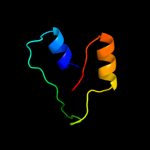
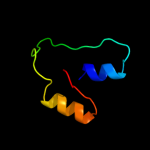
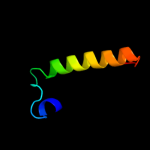
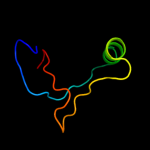
1 d1cmca_
100.0
100
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Met repressor, MetJ (MetR)2 c1is7F_
35.6
20
PDB header: hydrolase/protein bindingChain: F: PDB Molecule: gtp cyclohydrolase i;PDBTitle: crystal structure of rat gtpchi/gfrp stimulatory complex
3 d1wpla_
35.6
20
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: GTP cyclohydrolase I4 d1n3la_
27.2
21
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain5 d2pu9b1
25.6
45
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Ferredoxin thioredoxin reductase (FTR), alpha (variable) chain6 c2fekA_
25.6
15
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
7 c2wmyH_
24.1
13
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
8 c1q11A_
18.3
21
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: crystal structure of an active fragment of human tyrosyl-trna2 synthetase with tyrosinol
9 d2if6a1
17.9
32
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like10 c2j5bA_
16.1
20
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: structure of the tyrosyl trna synthetase from acanthamoeba2 polyphaga mimivirus complexed with tyrosynol
11 c2k29A_
14.7
24
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
12 d1txla_
14.5
31
Fold: LipocalinsSuperfamily: LipocalinsFamily: Hypothetical protein YodA13 c1txlA_
14.5
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: metal-binding protein yoda;PDBTitle: crystal structure of metal-binding protein yoda from e.2 coli, pfam duf149
14 c2y69Q_
14.5
18
PDB header: electron transportChain: Q: PDB Molecule: cytochrome c oxidase subunit 4 isoform 1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
15 d1q8da_
13.5
29
Fold: GDNF receptor-likeSuperfamily: GDNF receptor-likeFamily: GDNF receptor-like16 d2d5ra1
13.5
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: CAF1-like ribonuclease17 c2dlcX_
13.1
24
PDB header: ligase/trnaChain: X: PDB Molecule: tyrosyl-trna synthetase, cytoplasmic;PDBTitle: crystal structure of the ternary complex of yeast tyrosyl-trna2 synthetase
18 d1v54d_
12.6
18
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit IVFamily: Mitochondrial cytochrome c oxidase subunit IV19 d1qcsa2
11.2
17
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like20 d2bj7a1
10.6
21
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like21 d1a8ra_
not modelled
9.6
22
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: GTP cyclohydrolase I22 d1ioka2
not modelled
9.2
44
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain23 c3idwA_
not modelled
9.1
25
PDB header: endocytosisChain: A: PDB Molecule: actin cytoskeleton-regulatory complex protein sla1;PDBTitle: crystal structure of sla1 homology domain 2
24 c2v5eA_
not modelled
8.7
29
PDB header: receptor/glycoprotein complexChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: the structure of the gdnf\:coreceptor complex\: insights2 into ret signalling and heparin binding.
25 c3fubA_
not modelled
8.6
29
PDB header: hormoneChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: crystal structure of gdnf-gfralpha1 complex
26 d2hafa1
not modelled
8.5
27
Fold: VC0467-likeSuperfamily: VC0467-likeFamily: VC0467-like27 d2gf5a1
not modelled
8.4
17
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD28 c1qdnA_
not modelled
8.3
17
PDB header: fusion proteinChain: A: PDB Molecule: protein (n-ethylmaleimide sensitive fusionPDBTitle: amino terminal domain of the n-ethylmaleimide sensitive2 fusion protein (nsf)
29 c2cycB_
not modelled
8.2
18
PDB header: ligaseChain: B: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: crystal structure of tyrosyl-trna synthetase complexed with l-tyrosine2 from pyrococcus horikoshii
30 d1jf8a_
not modelled
8.2
16
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases31 c2l18A_
not modelled
7.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: an arsenate reductase in the phosphate binding state
32 d1sjpa2
not modelled
7.8
44
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain33 d1h8ba_
not modelled
7.7
21
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins34 d1sbxa_
not modelled
7.7
50
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Dachshund-homology domain35 d1wr6a1
not modelled
7.6
26
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain36 c3fwcN_
not modelled
7.3
23
PDB header: cell cycle, transcriptionChain: N: PDB Molecule: nuclear mrna export protein sac3;PDBTitle: sac3:sus1:cdc31 complex
37 d1dk7a_
not modelled
7.1
44
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain38 d1srva_
not modelled
7.1
56
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain39 d1oela2
not modelled
6.5
44
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain40 d1we3a2
not modelled
6.4
56
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain41 d1np3a1
not modelled
6.2
33
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Acetohydroxy acid isomeroreductase (ketol-acid reductoisomerase, KARI)42 d1xrsb2
not modelled
6.1
50
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain43 c2aj2A_
not modelled
6.0
27
PDB header: unknown functionChain: A: PDB Molecule: hypothetical upf0301 protein vc0467;PDBTitle: x-ray crystal structure of protein vc0467 from vibrio2 cholerae. northeast structural genomics consortium target3 vcr8.
44 c2p51A_
not modelled
5.9
32
PDB header: hydrolase, gene regulationChain: A: PDB Molecule: spcc18.06c protein;PDBTitle: crystal structure of the s. pombe pop2p deadenylation2 subunit
45 c1v91A_
not modelled
5.7
71
PDB header: toxinChain: A: PDB Molecule: delta-palutoxin it2;PDBTitle: solution structure of insectidal toxin delta-paluit2-nh2
46 d2dofa1
not modelled
5.7
13
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain47 d1wu2a2
not modelled
5.6
36
Fold: MoeA N-terminal region -likeSuperfamily: MoeA N-terminal region -likeFamily: MoeA N-terminal region -like48 c3t38B_
not modelled
5.5
33
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
49 d2bnma1
not modelled
5.4
29
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like50 c2cyaA_
not modelled
5.4
22
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: crystal structure of tyrosyl-trna synthetase from aeropyrum pernix
51 d1sjpa3
not modelled
5.3
0
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain52 d1wrda1
not modelled
5.2
17
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain53 d2cyua1
not modelled
5.2
11
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex54 d2nqra2
not modelled
5.0
36
Fold: MoeA N-terminal region -likeSuperfamily: MoeA N-terminal region -likeFamily: MoeA N-terminal region -like