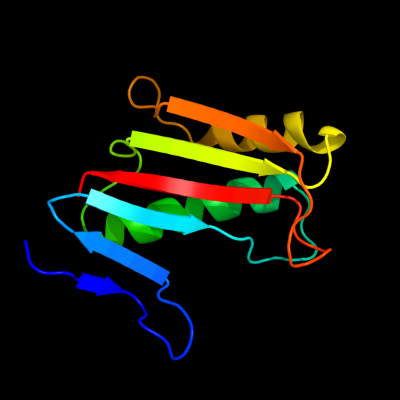
1 c3gtzA_
100.0
92
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative translation initiation inhibitor;PDBTitle: crystal structure of a putative translation initiation inhibitor from2 salmonella typhimurium
2 c3k12F_
100.0
35
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: uncharacterized protein a6v7t0;PDBTitle: crystal structure of an uncharacterized protein a6v7t0 from2 pseudomonas aeruginosa
3 c3kjjL_
100.0
43
PDB header: unknown functionChain: L: PDB Molecule: nmb1025 protein;PDBTitle: crystal structure of nmb1025, a member of yjgf protein family, from2 neisseria meningitidis (hexagonal crystal form)
4 d2ewca1
100.0
18
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP5 c3k0tA_
100.0
21
PDB header: sugar binding proteinChain: A: PDB Molecule: endoribonuclease l-psp, putative;PDBTitle: crystal structure of pspto -psp protein in complex with d-beta-glucose2 from pseudomonas syringae pv. tomato str. dc3000
6 d1jd1a_
100.0
26
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP7 c3m4sC_
100.0
26
PDB header: unknown functionChain: C: PDB Molecule: putative endoribonuclease l-psp;PDBTitle: crystal structure of a putative endoribonuclease l-psp from entamoeba2 histolytica, orthorhombic form
8 c3l7qD_
100.0
25
PDB header: translationChain: D: PDB Molecule: putative translation initiation inhibitor, aldr regulator-PDBTitle: crystal structure of aldr from streptococcus mutans
9 c3r0pB_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: l-psp putative endoribonuclease;PDBTitle: crystal structure of l-psp putative endoribonuclease from uncultured2 organism
10 c3lmeE_
100.0
17
PDB header: translationChain: E: PDB Molecule: possible translation initiation inhibitor;PDBTitle: structure of probable translation initiation inhibitor from2 (rpa2473) from rhodopseudomonas palustris
11 c3v4dC_
100.0
27
PDB header: oxidoreductaseChain: C: PDB Molecule: aminoacrylate peracid reductase rutc;PDBTitle: crystal structure of rutc protein a member of the yjgf family from2 e.coli
12 c3quwA_
100.0
23
PDB header: protein bindingChain: A: PDB Molecule: protein mmf1;PDBTitle: crystal structure of yeast mmf1
13 d2cvla1
100.0
28
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP14 d1onia_
100.0
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP15 d1nq3a_
100.0
26
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP16 c1xrgB_
100.0
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative translation initiation inhibitor, yjgfPDBTitle: conserved hypothetical protein from clostridium2 thermocellum cth-2968
17 d2b33a1
100.0
21
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP18 d1xrga_
100.0
23
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP19 d1qd9a_
100.0
31
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP20 d1x25a1
100.0
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP21 d1qaha_
not modelled
100.0
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP22 c3i7tA_
not modelled
100.0
18
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of rv2704, a member of highly conserved2 yjgf/yer057c/uk114 family, from mycobacterium tuberculosis
23 c2ig8C_
not modelled
100.0
27
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein pa3499;PDBTitle: crystal structure of a protein of unknown function pa3499 from2 pseudomonas aeruginosa
24 d1pf5a_
not modelled
100.0
21
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP25 d1j7ha_
not modelled
100.0
26
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP26 c2dyyG_
not modelled
100.0
26
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: upf0076 protein ph0854;PDBTitle: crystal structure of putative translation initiation2 inhibitor ph0854 from pyrococcus horikoshii
27 d2cwja1
not modelled
99.9
17
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP28 d2otma1
not modelled
99.9
14
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP29 d1qu9a_
not modelled
99.9
25
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP30 c3i3fB_
not modelled
99.9
25
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: hypothetical protein from giardia lamblia gl50803_14299
31 c3d01G_
not modelled
99.9
21
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the protein atu1372 with unknown function from2 agrobacterium tumefaciens
32 c3lybC_
not modelled
99.9
11
PDB header: hydrolaseChain: C: PDB Molecule: putative endoribonuclease;PDBTitle: structure of putative endoribonuclease(kp1_3112) from2 klebsiella pneumoniae
33 d1qnaa1
not modelled
56.8
7
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain34 d1mp9a1
not modelled
48.5
14
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain35 d1cdwa1
not modelled
45.6
11
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain36 d1nh2a1
not modelled
43.4
7
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain37 d1aisa1
not modelled
43.1
14
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain38 c1mp9B_
not modelled
29.1
16
PDB header: dna binding proteinChain: B: PDB Molecule: tata-binding protein;PDBTitle: tbp from a mesothermophilic archaeon, sulfolobus2 acidocaldarius
39 c2z8uQ_
not modelled
29.1
15
PDB header: transcriptionChain: Q: PDB Molecule: tata-box-binding protein;PDBTitle: methanococcus jannaschii tbp
40 c1d3uA_
not modelled
24.3
11
PDB header: gene regulation/dnaChain: A: PDB Molecule: tata-binding protein;PDBTitle: tata-binding protein/transcription factor (ii)b/bre+tata-2 box complex from pyrococcus woesei
41 d1cdwa2
not modelled
20.5
12
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain42 d1mp9a2
not modelled
19.1
10
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain43 d1qnaa2
not modelled
18.2
18
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain44 c1rm1A_
not modelled
17.3
11
PDB header: transcription/dnaChain: A: PDB Molecule: tata-box binding protein;PDBTitle: structure of a yeast tfiia/tbp/tata-box dna complex
45 c1ngmM_
not modelled
17.2
11
PDB header: transcription/dnaChain: M: PDB Molecule: transcription initiation factor tfiid;PDBTitle: crystal structure of a yeast brf1-tbp-dna ternary complex
46 d1nh2a2
not modelled
17.1
14
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain47 c3fqmA_
not modelled
15.5
20
PDB header: metal binding proteinChain: A: PDB Molecule: non-structural protein 5a;PDBTitle: crystal structure of a novel dimeric form of hcv ns5a domain i protein
48 d1aisa2
not modelled
15.0
15
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain49 c3eikB_
not modelled
14.0
14
PDB header: transcriptionChain: B: PDB Molecule: tata-box-binding protein;PDBTitle: double stranded dna binding protein
50 d1xhja_
not modelled
13.6
16
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like51 d1qh4a1
not modelled
13.5
13
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain52 d1g0wa1
not modelled
13.2
17
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain53 d1r5pa_
not modelled
12.6
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: KaiB-like54 d1u6ra1
not modelled
10.6
13
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain55 c3d87A_
not modelled
9.4
24
PDB header: cytokineChain: A: PDB Molecule: interleukin-23 subunit p19;PDBTitle: crystal structure of interleukin-23
56 c3hugA_
not modelled
9.2
6
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
57 d1vrpa1
not modelled
9.1
15
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain58 c3i4tA_
not modelled
9.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: diphthine synthase;PDBTitle: crystal structure of putative diphthine synthase from2 entamoeba histolytica
59 d1crka1
not modelled
8.7
17
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain60 d2qeda1
not modelled
8.2
17
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)61 d1i0ea1
not modelled
8.0
12
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain62 d2a6aa1
not modelled
8.0
3
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like63 d1nbwa2
not modelled
7.6
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ATPase domain of dehydratase reactivase alpha subunit64 c3mlcC_
not modelled
7.5
15
PDB header: isomeraseChain: C: PDB Molecule: fg41 malonate semialdehyde decarboxylase;PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
65 d1sxjc1
not modelled
7.1
8
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain66 d1xb2b1
not modelled
6.5
16
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain67 c1j4aA_
not modelled
6.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
68 d1qk1a1
not modelled
6.5
15
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain69 d2ix4a2
not modelled
6.2
11
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related70 c3a1yF_
not modelled
6.1
24
PDB header: ribosomal proteinChain: F: PDB Molecule: 50s ribosomal protein p1 (l12p);PDBTitle: the structure of protein complex
71 c1okjB_
not modelled
6.0
20
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protease yeaz;PDBTitle: crystal structure of the essential e. coli yeaz2 protein by mad method using the gadolinium complex3 "dotma"
72 d1qh5a_
not modelled
5.7
19
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)73 c2dvzA_
not modelled
5.7
10
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of a periplasmic transporter
74 d1y9ia_
not modelled
5.5
19
Fold: YutG-likeSuperfamily: YutG-likeFamily: YutG-like75 c2xd4A_
not modelled
5.5
10
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
76 c2qbbF_
not modelled
5.3
14
PDB header: ribosomeChain: F: PDB Molecule: 30s ribosomal protein s6;PDBTitle: crystal structure of the bacterial ribosome from2 escherichia coli in complex with gentamicin. this file3 contains the 30s subunit of the second 70s ribosome, with4 gentamicin bound. the entire crystal structure contains5 two 70s ribosomes and is described in remark 400.
77 d2qalf1
not modelled
5.3
14
Fold: Ferredoxin-likeSuperfamily: Ribosomal protein S6Family: Ribosomal protein S678 d1iqpa1
not modelled
5.3
21
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain