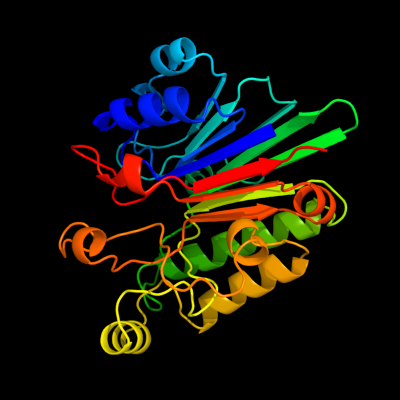
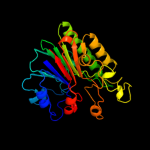
1 d1akoa_
100.0
100
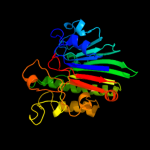
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like2 c2voaB_
100.0
31
PDB header: lyaseChain: B: PDB Molecule: exodeoxyribonuclease iii;PDBTitle: structure of an ap endonuclease from archaeoglobus fulgidus
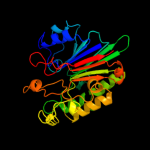
3 c2jc4A_
100.0
36
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease iii;PDBTitle: 3'-5' exonuclease (nexo) from neisseria meningitidis
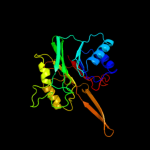
4 c2j63B_
100.0
23
PDB header: lyaseChain: B: PDB Molecule: ap-endonuclease;PDBTitle: crystal structure of ap endonuclease lmap from leishmania2 major
5 c3g0rA_
100.0
28
PDB header: hydrolase/dnaChain: A: PDB Molecule: exodeoxyribonuclease;PDBTitle: complex of mth0212 and an 8bp dsdna with distorted ends
6 c2jc5A_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease;PDBTitle: apurinic apyrimidinic (ap) endonuclease (nape) from2 neisseria meningitidis
7 c1e9nB_
100.0
24
PDB header: dna repairChain: B: PDB Molecule: dna-(apurinic or apyrimidinic site) lyase;PDBTitle: a second divalent metal ion in the active site of a new2 crystal form of human apurinic/apyrimidinic endonuclease,3 ape1, and its implications for the catalytic mechanism
8 c3tebA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease/exonuclease/phosphatase;PDBTitle: endonuclease/exonuclease/phosphatase family protein from leptotrichia2 buccalis c-1013-b
9 d1vyba_
100.0
24
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like10 d1hd7a_
100.0
25
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like11 c3g6sA_
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: putative endonuclease/exonuclease/phosphatasePDBTitle: crystal structure of the2 endonuclease/exonuclease/phosphatase (bvu_0621) from3 bacteroides vulgatus. northeast structural genomics4 consortium target bvr56d
12 c3ngoA_
100.0
16
PDB header: hydrolase/dnaChain: A: PDB Molecule: ccr4-not transcription complex subunit 6-like;PDBTitle: crystal structure of the human cnot6l nuclease domain in complex with2 poly(a) dna
13 c3mprB_
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: putative endonuclease/exonuclease/phosphatase familyPDBTitle: crystal structure of endonuclease/exonuclease/phosphatase family2 protein from bacteroides thetaiotaomicron, northeast structural3 genomics consortium target btr318a
14 d2imqx1
100.0
13
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)15 d1zwxa1
100.0
14
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Sphingomyelin phosphodiesterase-like16 c3i46B_
99.9
17
PDB header: toxinChain: B: PDB Molecule: beta-hemolysin;PDBTitle: crystal structure of beta toxin from staphylococcus aureus f277a,2 p278a mutant with bound calcium ions
17 c3mtcA_
99.9
14
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: type ii inositol-1,4,5-trisphosphate 5-phosphatase;PDBTitle: crystal structure of inpp5b in complex with phosphatidylinositol 4-2 phosphate
18 c3l1wE_
99.9
16
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of a functionally unknown conserved2 protein from enterococcus faecalis v583
19 d2ddra1
99.9
20
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Sphingomyelin phosphodiesterase-like20 c3nr8A_
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2;PDBTitle: crystal structure of human ship2
21 d1wdua_
not modelled
99.9
15
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like22 d1sr4b_
not modelled
99.9
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like23 d2f1na1
not modelled
99.9
20
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like24 c2ei9A_
not modelled
99.9
14
PDB header: gene regulationChain: A: PDB Molecule: non-ltr retrotransposon r1bmks orf2 protein;PDBTitle: crystal structure of r1bm endonuclease domain
25 d2a40b1
not modelled
99.9
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like26 c2xswB_
not modelled
99.8
16
PDB header: hydrolaseChain: B: PDB Molecule: 72 kda inositol polyphosphate 5-phosphatase;PDBTitle: crystal structure of human inpp5e
27 d1i9za_
not modelled
99.8
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)28 d3bula2
not modelled
46.3
22
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain29 c2vhiG_
not modelled
43.2
8
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
30 c2i2xD_
not modelled
24.3
22
PDB header: transferaseChain: D: PDB Molecule: methyltransferase 1;PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
31 d1xrsb1
not modelled
23.4
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain32 d1f89a_
not modelled
19.9
15
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase33 d1emsa2
not modelled
19.5
6
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase34 c1xrsB_
not modelled
17.3
13
PDB header: isomeraseChain: B: PDB Molecule: d-lysine 5,6-aminomutase beta subunit;PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
35 c2yxbA_
not modelled
16.9
37
PDB header: isomeraseChain: A: PDB Molecule: coenzyme b12-dependent mutase;PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
36 c1y80A_
not modelled
16.6
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted cobalamin binding protein;PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
37 d1ccwa_
not modelled
16.1
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain38 c1bmtB_
not modelled
15.7
22
PDB header: methyltransferaseChain: B: PDB Molecule: methionine synthase;PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
39 c2gm2A_
not modelled
14.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
40 c1k98A_
not modelled
13.8
22
PDB header: transferaseChain: A: PDB Molecule: methionine synthase;PDBTitle: adomet complex of meth c-terminal fragment
41 c3jvvA_
not modelled
12.5
15
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
42 d1g6oa_
not modelled
11.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)43 c2w1vA_
not modelled
10.8
22
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
44 d1uf5a_
not modelled
10.2
14
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase45 c3ezxA_
not modelled
9.8
17
PDB header: transferaseChain: A: PDB Molecule: monomethylamine corrinoid protein 1;PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
46 c3hkxA_
not modelled
9.6
9
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
47 c3nrbD_
not modelled
9.4
41
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
48 c3n0vD_
not modelled
9.3
12
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
49 c2eyuA_
not modelled
8.9
16
PDB header: protein transportChain: A: PDB Molecule: twitching motility protein pilt;PDBTitle: the crystal structure of the c-terminal domain of aquifex2 aeolicus pilt
50 c3n05B_
not modelled
8.7
6
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
51 c2p10D_
not modelled
8.5
15
PDB header: hydrolaseChain: D: PDB Molecule: mll9387 protein;PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
52 c3o1lB_
not modelled
8.1
35
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
53 c3obiC_
not modelled
8.1
18
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
54 c2gzaB_
not modelled
8.1
16
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein virb11;PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
55 c3louB_
not modelled
7.9
18
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
56 d1p9ra_
not modelled
7.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)57 c1emsB_
not modelled
7.3
9
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
58 c2dclB_
not modelled
7.1
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical upf0166 protein ph1503;PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
59 d1j31a_
not modelled
6.9
18
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase60 c3p9xB_
not modelled
6.9
18
PDB header: transferaseChain: B: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
61 c3q9cF_
not modelled
6.8
7
PDB header: hydrolaseChain: F: PDB Molecule: acetylpolyamine amidohydrolase;PDBTitle: crystal structure of h159a apah complexed with n8-acetylspermidine
62 d2odka1
not modelled
6.6
12
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like63 d3bzka5
not modelled
6.4
33
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like64 c3kxyB_
not modelled
6.4
13
PDB header: chaperone/transcription inhibitorChain: B: PDB Molecule: exoenzyme s synthesis protein c;PDBTitle: crystal structure of the exsc-exse complex
65 d1fmfa_
not modelled
6.3
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain66 d2fvta1
not modelled
6.3
6
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like67 d2eg6a1
not modelled
6.2
50
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Dihydroorotase68 d1zgha2
not modelled
6.0
15
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase69 c2odkD_
not modelled
5.9
11
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
70 d1noya_
not modelled
5.9
12
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease71 d1ih7a1
not modelled
5.9
6
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease72 d1o51a_
not modelled
5.7
8
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: DUF190/COG199373 c3jzeC_
not modelled
5.6
50
PDB header: hydrolaseChain: C: PDB Molecule: dihydroorotase;PDBTitle: 1.8 angstrom resolution crystal structure of dihydroorotase (pyrc)2 from salmonella enterica subsp. enterica serovar typhimurium str. lt2
74 c2gszE_
not modelled
5.5
21
PDB header: protein transportChain: E: PDB Molecule: twitching motility protein pilt;PDBTitle: structure of a. aeolicus pilt with 6 monomers per2 asymmetric unit
75 c3pnuA_
not modelled
5.4
67
PDB header: hydrolaseChain: A: PDB Molecule: dihydroorotase;PDBTitle: 2.4 angstrom crystal structure of dihydroorotase (pyrc) from2 campylobacter jejuni.
76 d1jkxa_
not modelled
5.2
29
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase77 d1uf3a_
not modelled
5.2
17
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like