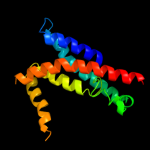
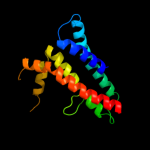
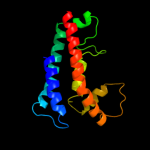
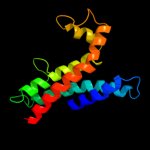
1 d1kqfc_
99.9
16
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Formate dehydrogenase N, cytochrome (gamma) subunit2 c2qjkM_
95.5
15
PDB header: electron transportChain: M: PDB Molecule: cytochrome b;PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
3 d3cx5c2
95.1
16
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)4 c3cx5N_
94.5
16
PDB header: oxidoreductaseChain: N: PDB Molecule: cytochrome b;PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
5 c3cwbC_
94.0
16
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome b;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
6 d2e74a1
92.7
15
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)7 d1ppjc2
91.8
16
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)8 d1q90b_
91.1
15
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)9 d1bccc3
90.0
16
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)10 d1y5ic1
81.3
11
Fold: Heme-binding four-helical bundleSuperfamily: Respiratory nitrate reductase 1 gamma chainFamily: Respiratory nitrate reductase 1 gamma chain11 c2ftcQ_
23.0
36
PDB header: ribosomeChain: Q: PDB Molecule: 39s ribosomal protein l34, mitochondrial;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
12 d1t33a1
15.8
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain13 d2ieca1
12.6
20
Fold: MK0786-likeSuperfamily: MK0786-likeFamily: MK0786-like14 d1m56d_
10.5
14
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV15 c3nnrA_
10.4
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of a tetr-family transcriptional regulator2 (maqu_3571) from marinobacter aquaeolei vt8 at 2.49 a resolution
16 c2ogfD_
10.3
30
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein mj0408;PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
17 d2id6a1
10.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain18 c2yvhA_
10.1
22
PDB header: transcription/dnaChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of the operator-binding form of the multi-drug2 binding transcriptional repressor cgmr
19 c2raeA_
9.8
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, acrr family protein;PDBTitle: crystal structure of a tetr/acrr family transcriptional regulator from2 rhodococcus sp. rha1
20 d2i52a1
9.7
30
Fold: MK0786-likeSuperfamily: MK0786-likeFamily: MK0786-like21 c1z0xA_
not modelled
9.6
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of transcriptional regulator, tetr family from2 enterococcus faecalis v583
22 c2guhA_
not modelled
9.4
6
PDB header: transcriptionChain: A: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: crystal structure of the putative tetr-family transcriptional2 regulator from rhodococcus sp. rha1
23 d2d6ya1
not modelled
9.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain24 c2qibA_
not modelled
8.7
11
PDB header: transcriptionChain: A: PDB Molecule: tetr-family transcriptional regulator;PDBTitle: crystal structure of tetr-family transcriptional regulator from2 streptomyces coelicolor
25 d2gfna1
not modelled
8.5
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain26 c2dg8D_
not modelled
8.4
11
PDB header: gene regulationChain: D: PDB Molecule: putative tetr-family transcriptional regulatory protein;PDBTitle: crystal structure of the putative trasncriptional regulator sco75182 from streptomyces coelicolor a3(2)
27 d1z0xa1
not modelled
8.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain28 c2wgbB_
not modelled
7.7
11
PDB header: transcriptionChain: B: PDB Molecule: tetr family transcriptional repressor lfrr;PDBTitle: crystal structure of the tetr-like transcriptional2 regulator lfrr from mycobacterium smegmatis
29 d1uf2c2
not modelled
7.6
33
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Top domain of virus capsid protein30 d2g3ba1
not modelled
7.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain31 c2hxiA_
not modelled
7.5
22
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structural genomics, the crystal structure of a putative2 transcriptional regulator from streptomyces coelicolor a3(2)
32 d2fx0a1
not modelled
7.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain33 d2vkea1
not modelled
7.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain34 c2fbqA_
not modelled
7.2
22
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional regulator;PDBTitle: the crystal structure of transcriptional regulator pa3006
35 d2j7ja2
not modelled
7.1
10
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H236 d2o7ta1
not modelled
7.1
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain37 d2hkua1
not modelled
6.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain38 d2id3a1
not modelled
6.6
0
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain39 c3fiwB_
not modelled
6.5
17
PDB header: transcription regulatorChain: B: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: structure of sco0253, a tetr-family transcriptional regulator from2 streptomyces coelicolor
40 d1vi0a1
not modelled
6.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain41 d2g7ga1
not modelled
6.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain42 d2vkva1
not modelled
6.2
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain43 c2g7sA_
not modelled
6.2
22
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: the crystal structure of transcriptional regulator, tetr family, from2 agrobacterium tumefaciens
44 d1v7ba1
not modelled
6.1
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain45 c3efdK_
not modelled
6.1
33
PDB header: immune systemChain: K: PDB Molecule: kcsa;PDBTitle: the crystal structure of the cytoplasmic domain of kcsa
46 c2id3A_
not modelled
6.1
0
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of transcriptional regulator sco5951 from2 streptomyces coelicolor a3(2)
47 d2i10a1
not modelled
6.1
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain48 d1qled_
not modelled
6.1
7
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV49 c3c2bA_
not modelled
6.0
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of tetr transcriptional regulator from agrobacterium2 tumefaciens
50 d2axti1
not modelled
6.0
14
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like51 c3rh2A_
not modelled
6.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: hypothetical tetr-like transcriptional regulator;PDBTitle: crystal structure of a hypothetical tetr-like transcriptional2 regulator (sama_0099) from shewanella amazonensis sb2b at 2.42 a3 resolution
52 c2g3bB_
not modelled
5.9
17
PDB header: transcriptionChain: B: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: crystal structure of putative tetr-family transcriptional regulator2 from rhodococcus sp.
53 c3cwrA_
not modelled
5.9
6
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of transcriptional regulator of tetr2 family (yp_425770.1) from rhodospirillum rubrum atcc 111703 at 1.50 a resolution
54 c3bniA_
not modelled
5.9
6
PDB header: transcription regulatorChain: A: PDB Molecule: putative tetr-family transcriptional regulator;PDBTitle: crystal structure of tetr-family transcriptional regulator from2 streptomyces coelicolor
55 d2axtb1
not modelled
5.8
22
Fold: Photosystem II antenna protein-likeSuperfamily: Photosystem II antenna protein-likeFamily: Photosystem II antenna protein-like56 d2axtc1
not modelled
5.8
22
Fold: Photosystem II antenna protein-likeSuperfamily: Photosystem II antenna protein-likeFamily: Photosystem II antenna protein-like57 c2axtc_
not modelled
5.8
22
PDB header: electron transportChain: C: PDB Molecule: photosystem ii cp43 protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
58 c2d6yA_
not modelled
5.8
11
PDB header: gene regulationChain: A: PDB Molecule: putative tetr family regulatory protein;PDBTitle: crystal structure of transcriptional factor sco4008 from streptomyces2 coelicolor a3(2)
59 c2zcxA_
not modelled
5.7
17
PDB header: transcriptionChain: A: PDB Molecule: tetr-family transcriptional regulator;PDBTitle: crystal structure of tetr family transcriptional regulator2 sco7815
60 c2dg7A_
not modelled
5.5
11
PDB header: gene regulationChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of the putative transcriptional regulator sco03372 from streptomyces coelicolor a3(2)
61 c2o7tA_
not modelled
5.4
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of a tetr family transcriptional regulator2 (ncgl1578, cgl1640) from corynebacterium glutamicum at 2.10 a3 resolution
62 d2g7sa1
not modelled
5.4
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain63 c1t33B_
not modelled
5.4
22
PDB header: transcriptionChain: B: PDB Molecule: putative transcriptional repressor (tetr/acrr family);PDBTitle: structural genomics, the crystal structure of a putative2 transcriptional repressor (tetr/acrr family) from salmonella3 typhimurim lt2
64 d1pb6a1
not modelled
5.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain65 c3o60A_
not modelled
5.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin0861 protein;PDBTitle: the crystal structure of lin0861 from listeria innocua to 2.8a
66 c3egqB_
not modelled
5.2
17
PDB header: transcriptionChain: B: PDB Molecule: tetr family transcriptional regulator;PDBTitle: crystal structure of a tetr-family transcriptional regulator (af_1817)2 from archaeoglobus fulgidus at 2.55 a resolution
67 d1jt6a1
not modelled
5.2
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain