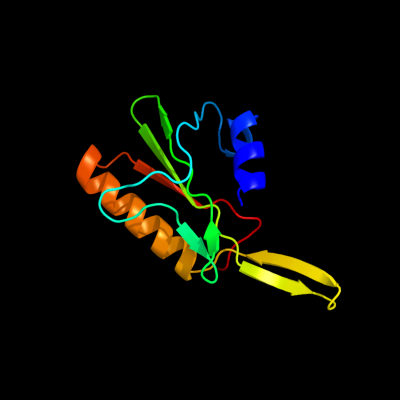
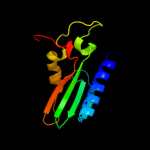
1 c2vldA_
88.9
20
PDB header: hydrolaseChain: A: PDB Molecule: upf0286 protein pyrab01260;PDBTitle: crystal structure of a repair endonuclease from pyrococcus2 abyssi
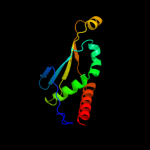
2 d1xmxa_
70.7
13
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hypothetical protein VC18993 d2foka4
69.0
17
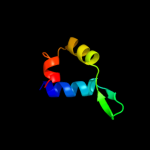
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease FokI, C-terminal (catalytic) domain4 c2wj0B_
64.5
18
PDB header: hydrolase/dnaChain: B: PDB Molecule: archaeal hjc;PDBTitle: crystal structures of holliday junction resolvases from2 archaeoglobus fulgidus bound to dna substrate
5 c1fokA_
62.2
17
PDB header: hydrolase/dnaChain: A: PDB Molecule: protein (foki restriction endonucleas);PDBTitle: structure of restriction endonuclease foki bound to dna
6 c3a9fA_
56.6
12
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crystal structure of the c-terminal domain of cytochrome cz2 from chlorobium tepidum
7 c1y88A_
52.7
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein af1548;PDBTitle: crystal structure of protein of unknown function af1548
8 d1hh1a_
50.5
14
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like9 d1ob8a_
39.4
16
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like10 d1y88a2
36.7
6
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: MRR-like11 d1gefa_
33.5
14
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like12 c2fhyL_
32.5
21
PDB header: hydrolaseChain: L: PDB Molecule: fructose-1,6-bisphosphatase 1;PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
13 d1spia_
30.1
21
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like14 d1nuwa_
28.9
25
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like15 c2gq1A_
28.8
18
PDB header: hydrolaseChain: A: PDB Molecule: fructose-1,6-bisphosphatase;PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
16 d1bk4a_
28.6
21
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like17 d1ftaa_
28.2
21
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like18 c2e52A_
21.5
14
PDB header: hydrolase/dnaChain: A: PDB Molecule: type ii restriction enzyme hindiii;PDBTitle: crystal structural analysis of hindiii restriction endonuclease in2 complex with cognate dna at 2.0 angstrom resolution
19 c3h4rA_
17.9
16
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease 8;PDBTitle: crystal structure of e. coli rece exonuclease
20 c2l2oA_
15.4
6
PDB header: unknown functionChain: A: PDB Molecule: upf0727 protein c6orf115;PDBTitle: solution structure of human hspc280 protein
21 c3h1tA_
not modelled
12.9
22
PDB header: hydrolaseChain: A: PDB Molecule: type i site-specific restriction-modificationPDBTitle: the fragment structure of a putative hsdr subunit of a type2 i restriction enzyme from vibrio vulnificus yj016
22 d1c3ha_
not modelled
12.5
30
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like23 d1d9qa_
not modelled
11.9
22
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like24 c2krhA_
not modelled
10.8
9
PDB header: actin-binding proteinChain: A: PDB Molecule: actin-binding rho-activating protein;PDBTitle: structure of the c-terminal actin binding domain of abra
25 d2bu3a1
not modelled
7.9
12
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Phytochelatin synthase26 d1w36b3
not modelled
5.9
16
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Exodeoxyribonuclease V beta chain (RecB), C-terminal domain27 d1pk6a_
not modelled
5.5
50
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like