|
|
 |
| Summary |
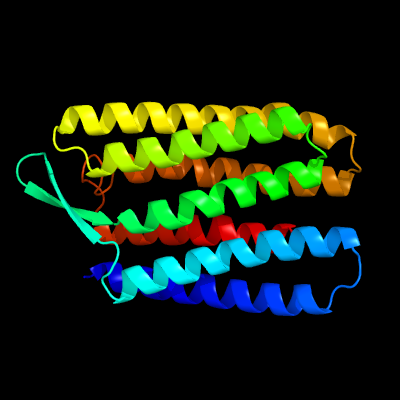
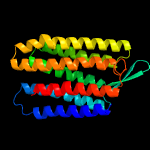
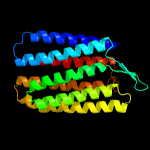
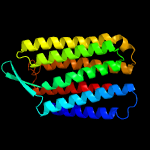
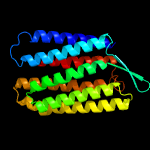
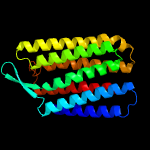
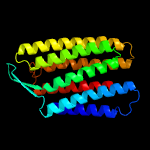
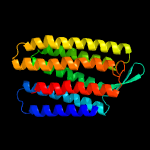
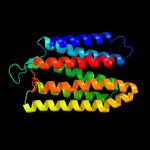
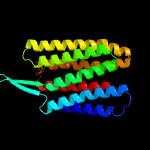
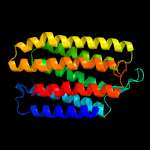
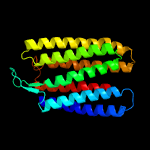
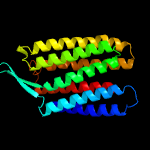
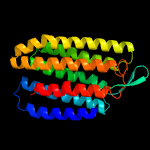
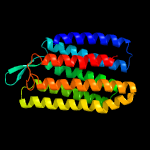
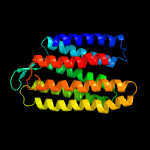
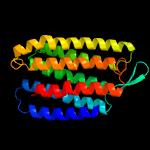
| Top model | |||||||||||||||
| |||||||||||||||
| Sequence analysis |
| Download FASTA version |
| Secondary structure and disorder prediction [Show] |
 |
| 1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 | |||||||||||||||||||||
| Sequence | R | P | E | W | I | W | L | A | L | G | T | A | L | M | G | L | G | T | L | Y | F | L | V | K | G | M | G | V | S | D | P | D | A | K | K | F | Y | A | I | T | T | L | V | P | A | I | A | F | T | M | Y | L | S | M | L | L | G | Y | G | L | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | |||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 | |||||||||||||||||||||
| Sequence | T | M | V | P | F | G | G | E | Q | N | P | I | Y | W | A | R | Y | A | D | W | L | F | T | T | P | L | L | L | L | D | L | A | L | L | V | D | A | D | Q | G | T | I | L | A | L | V | G | A | D | G | I | M | I | G | T | G | L | V | G | A | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . | . | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | . | . | 180 | |||||||||||||||||||||
| Sequence | L | T | K | V | Y | S | Y | R | F | V | W | W | A | I | S | T | A | A | M | L | Y | I | L | Y | V | L | F | F | G | F | T | S | K | A | E | S | M | R | P | E | V | A | S | T | F | K | V | L | R | N | V | T | V | V | L | W | S | A | Y | P | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 190 | . | . | . | . | . | . | . | . | . | 200 | . | . | . | . | . | . | . | . | . | 210 | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||||||||||||||||||
| Sequence | V | V | W | L | I | G | S | E | G | A | G | I | V | P | L | N | I | E | T | L | L | F | M | V | L | D | V | S | A | K | V | G | F | G | L | I | L | L | R | |||||||||||||||||||||||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||
| Domain analysis [Show] |
 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| Detailed template information [Hide] |
| Binding site prediction |
Automated 3DLigandsite submission is temporarily suspended due to server load. However you may submit models directly to 3DLigandSite HERE
| Transmembrane helix prediction |
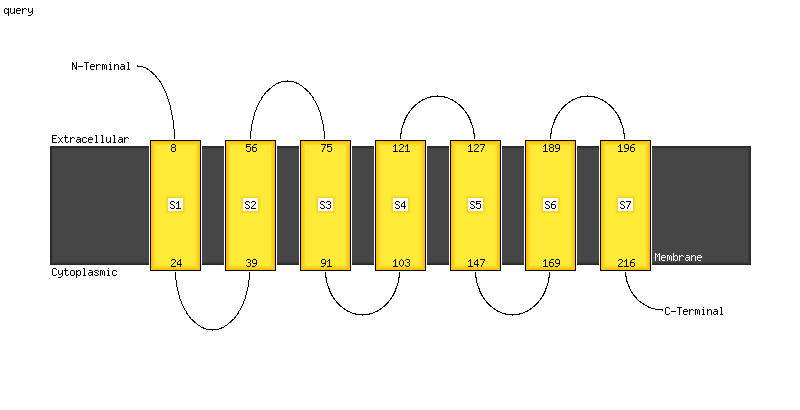
Transmembrane helices have been predicted in your sequence to adopt the topology shown below
Phyre is now FREE for commercial users!
All images and data generated by Phyre2 are free to use in any publication with acknowledgement
Accessibility Statement| Please cite: Phyre2.2: A Community Resource for Template-based Protein Structure Prediction | |||||||||
| Powell HR et al. Journal of Molecular Biology (2025) in press DOI: https://doi.org/10.1016/j.jmb.2025.168960 | |||||||||
|
 |
|
|||||||