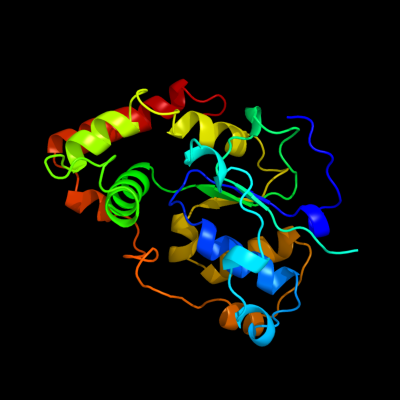
1 d1fmja_
100.0
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase2 d1q44a_
100.0
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase3 c2h8kA_
100.0
20
PDB header: transferaseChain: A: PDB Molecule: sult1c3 splice variant d;PDBTitle: human sulfotranferase sult1c3 in complex with pap
4 d1q20a_
100.0
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase5 d1g3ma_
100.0
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase6 d1xv1a_
100.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase7 c2zvpX_
100.0
18
PDB header: transferaseChain: X: PDB Molecule: tyrosine-ester sulfotransferase;PDBTitle: crystal structure of mouse cytosolic sulfotransferase msult1d1 complex2 with pap and p-nitrophenol
8 d1ls6a_
100.0
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase9 c3u3oA_
100.0
15
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase 1a1;PDBTitle: crystal structure of human sult1a1 bound to pap and two 3-cyano-7-2 hydroxycoumarin
10 d1aqua_
100.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase11 d2a3ra1
100.0
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase12 c1q1qA_
100.0
19
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase family, cytosolic, 2b, member 1PDBTitle: crystal structure of human pregnenolone sulfotransferase2 (sult2b1a) in the presence of pap
13 c1zd1B_
100.0
17
PDB header: transferaseChain: B: PDB Molecule: sulfotransferase 4a1;PDBTitle: human sulfortransferase sult4a1
14 d1j99a_
100.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase15 d3bfxa1
100.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase16 c2gwhA_
100.0
18
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase 1c2;PDBTitle: human sulfotranferase sult1c2 in complex with pap and2 pentachlorophenol
17 c3mgbA_
100.0
18
PDB header: transferase/antibioticChain: A: PDB Molecule: teg12;PDBTitle: teg 12 ternary structure complexed with pap and the teicoplanin2 aglycone
18 c3mg9A_
100.0
18
PDB header: transferase/antibioticChain: A: PDB Molecule: teg12;PDBTitle: teg 12 binary structure complexed with the teicoplanin aglycone
19 c2ovfA_
100.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: stal;PDBTitle: crystal structure of stal-pap complex
20 c3ap3A_
99.9
16
PDB header: transferaseChain: A: PDB Molecule: protein-tyrosine sulfotransferase 2;PDBTitle: crystal structure of human tyrosylprotein sulfotransferase-2 complexed2 with pap
21 d1t8ta_
not modelled
99.9
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase22 c1vkjA_
not modelled
99.9
13
PDB header: transferaseChain: A: PDB Molecule: heparan sulfate (glucosamine) 3-o-PDBTitle: crystal structure of heparan sulfate 3-o-sulfotransferase2 isoform 1 in the presence of pap
23 d1vkja_
not modelled
99.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase24 c1nstA_
not modelled
99.9
15
PDB header: sulfotransferaseChain: A: PDB Molecule: heparan sulfate n-deacetylase/n-sulfotransferase;PDBTitle: the sulfotransferase domain of human haparin sulfate n-2 deacetylase/n-sulfotransferase
25 d1nsta_
not modelled
99.9
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase26 c3rnlA_
not modelled
99.8
14
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase;PDBTitle: crystal structure of sulfotransferase from alicyclobacillus2 acidocaldarius
27 c3bd9A_
not modelled
99.8
10
PDB header: transferaseChain: A: PDB Molecule: heparan sulfate glucosamine 3-o-sulfotransferasePDBTitle: human 3-o-sulfotransferase isoform 5 with bound pap
28 c2z6vA_
not modelled
99.7
14
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of sulfotransferase stf9 from2 mycobacterium avium
29 c2zq5A_
not modelled
99.6
12
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of sulfotransferase stf1 from2 mycobacterium tuberculosis h37rv (type1 form)
30 d1texa_
not modelled
99.5
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase31 c3f5fA_
not modelled
94.7
15
PDB header: transport, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein, heparanPDBTitle: crystal structure of heparan sulfate 2-o-sulfotransferase2 from gallus gallus as a maltose binding protein fusion.
32 d2cbia2
not modelled
64.0
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain33 c1nijA_
not modelled
52.7
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yjia;PDBTitle: yjia protein
34 c2xsbA_
not modelled
48.2
23
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: ogoga pugnac complex
35 d1qgva_
not modelled
45.6
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: spliceosomal protein U5-15Kd36 d1nija1
not modelled
41.8
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like37 d1s2xa_
not modelled
37.0
25
Fold: STAT-likeSuperfamily: Cag-ZFamily: Cag-Z38 c1s2xA_
not modelled
37.0
25
PDB header: unknown functionChain: A: PDB Molecule: cag-z;PDBTitle: crystal structure of cag-z from helicobacter pylori
39 d2choa2
not modelled
36.8
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain40 c1ti5A_
not modelled
35.9
27
PDB header: plant proteinChain: A: PDB Molecule: plant defensin;PDBTitle: solution structure of plant defensin
41 d1jkza_
not modelled
35.1
24
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Plant defensins42 c2cbjA_
not modelled
28.1
23
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronidase;PDBTitle: structure of the clostridium perfringens nagj family 842 glycoside hydrolase, a homologue of human o-glcnacase in3 complex with pugnac
43 c1yj5B_
not modelled
27.6
22
PDB header: transferaseChain: B: PDB Molecule: 5' polynucleotide kinase-3' phosphatase catalytic domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
44 c3zvmA_
not modelled
24.7
27
PDB header: hydrolase/transferase/dnaChain: A: PDB Molecule: bifunctional polynucleotide phosphatase/kinase;PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
45 d1wiza_
not modelled
24.0
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain46 d1j09a2
not modelled
22.4
13
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain47 c3c52B_
not modelled
21.2
23
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
48 c2j8lA_
not modelled
19.8
15
PDB header: hydrolaseChain: A: PDB Molecule: coagulation factor xi;PDBTitle: fxi apple 4 domain loop-out conformation
49 d1dx5i3
not modelled
18.9
26
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-type module50 d1l3ya_
not modelled
18.5
41
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: Integrin beta EGF-like domains51 c1wxnA_
not modelled
17.6
46
PDB header: toxinChain: A: PDB Molecule: toxin apetx2;PDBTitle: solution structure of apetx2, a specific peptide inhibitor2 of asic3 proton-gated channels
52 d1ppjd1
not modelled
16.3
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain53 d1rz3a_
not modelled
16.3
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase54 d1pvza_
not modelled
15.4
27
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins55 c2j8jB_
not modelled
15.2
19
PDB header: hydrolaseChain: B: PDB Molecule: coagulation factor xi;PDBTitle: solution structure of the a4 domain of blood coagulation2 factor xi
56 c1wqkA_
not modelled
14.2
55
PDB header: toxinChain: A: PDB Molecule: toxin apetx1;PDBTitle: solution structure of apetx1, a specific peptide inhibitor2 of human ether-a-go-go-related gene potassium channels3 from the venom of the sea anemone anthopleura4 elegantissima: a new fold for an herg toxin
57 c2j8lB_
not modelled
12.5
19
PDB header: hydrolaseChain: B: PDB Molecule: coagulation factor xi;PDBTitle: fxi apple 4 domain loop-out conformation
58 c2choA_
not modelled
12.5
15
PDB header: hydrolaseChain: A: PDB Molecule: glucosaminidase;PDBTitle: bacteroides thetaiotaomicron hexosaminidase with o-2 glcnacase activity
59 c2j8jA_
not modelled
12.1
15
PDB header: hydrolaseChain: A: PDB Molecule: coagulation factor xi;PDBTitle: solution structure of the a4 domain of blood coagulation2 factor xi
60 d1cfza_
not modelled
11.4
17
Fold: Phosphorylase/hydrolase-likeSuperfamily: HybD-likeFamily: Hydrogenase maturating endopeptidase HybD61 d1esfa1
not modelled
11.0
25
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain62 c3pesA_
not modelled
11.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein gp49;PDBTitle: crystal structure of uncharacterized protein from pseudomonas phage2 yua
63 d1ekga_
not modelled
10.8
31
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like64 c3cwbQ_
not modelled
10.4
34
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
65 d1tafb_
not modelled
10.2
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs66 c1p84D_
not modelled
10.2
17
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
67 d1s6ya2
not modelled
10.2
17
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase68 c1ly1A_
not modelled
9.8
43
PDB header: transferaseChain: A: PDB Molecule: polynucleotide kinase;PDBTitle: structure and mechanism of t4 polynucleotide kinase
69 d1ly1a_
not modelled
9.8
43
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases70 c2kp0A_
not modelled
9.2
22
PDB header: de novo proteinChain: A: PDB Molecule: nasonin-1m;PDBTitle: solution structure of nasonin-1m
71 c3izcw_
not modelled
9.1
15
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein rpl22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
72 c2w1oA_
not modelled
8.7
23
PDB header: translationChain: A: PDB Molecule: 60s acidic ribosomal protein p2;PDBTitle: nmr structure of dimerization domain of human ribosomal2 protein p2
73 d1h9ii_
not modelled
8.5
25
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Plant inhibitors of proteinases and amylasesFamily: Plant inhibitors of proteinases and amylases74 c2a45L_
not modelled
8.1
38
PDB header: hydrolase/hydrolase inhibitorChain: L: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
75 d1ozza_
not modelled
8.1
29
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Insect defensins76 c1yseA_
not modelled
8.0
20
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: solution structure of the mar-binding domain of satb1
77 c3iz5w_
not modelled
7.9
19
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
78 c3enoC_
not modelled
7.9
29
PDB header: hydrolase/unknown functionChain: C: PDB Molecule: uncharacterized protein pf2011;PDBTitle: crystal structure of pyrococcus furiosus pcc1 in complex2 with thermoplasma acidophilum kae1
79 d1iwea_
not modelled
7.9
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like80 c1xbsA_
not modelled
7.9
20
PDB header: transcription, cell cycleChain: A: PDB Molecule: dim1-like protein;PDBTitle: crystal structure of human dim2: a dim1-like protein
81 c2c1eB_
not modelled
7.8
30
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: caspase-3 subunit p12;PDBTitle: crystal structures of caspase-3 in complex with aza-peptide michael2 acceptor inhibitors.
82 d1yj5a2
not modelled
7.8
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases83 c2f1rA_
not modelled
7.8
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesisPDBTitle: crystal structure of molybdopterin-guanine biosynthesis2 protein b (mobb)
84 d1myna_
not modelled
7.8
23
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Insect defensins85 c3bs3A_
not modelled
7.7
23
PDB header: dna binding proteinChain: A: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of a putative dna-binding protein from bacteroides2 fragilis
86 c3dnlG_
not modelled
7.2
25
PDB header: viral proteinChain: G: PDB Molecule: hiv-1 envelope glycoprotein gp120;PDBTitle: molecular structure for the hiv-1 gp120 trimer in the b12-2 bound state
87 d1sisa_
not modelled
7.1
38
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins88 d1q90a3
not modelled
6.8
16
Fold: Single transmembrane helixSuperfamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchorFamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchor89 d2ga5a1
not modelled
6.8
38
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like90 c2e3eA_
not modelled
6.8
17
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin, mutant def-bbb;PDBTitle: nmr structure of def-bbb, a mutant of anopheles defensin2 def-aaa
91 c2kz5A_
not modelled
6.7
18
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
92 d1m8pa3
not modelled
6.6
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain93 c1iweB_
not modelled
6.5
28
PDB header: ligaseChain: B: PDB Molecule: adenylosuccinate synthetase;PDBTitle: imp complex of the recombinant mouse-muscle2 adenylosuccinate synthetase
94 d1lmb3_
not modelled
6.5
11
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors95 c3b85A_
not modelled
6.4
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphate starvation-inducible protein;PDBTitle: crystal structure of predicted phosphate starvation-induced atpase2 phoh2 from corynebacterium glutamicum
96 c2nz3A_
not modelled
6.4
30
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin, mutant def-acaa;PDBTitle: nmr structure of def-acaa, a mutant of anopheles defensin2 def-aaa
97 d1icaa_
not modelled
6.3
26
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Insect defensins98 d1zela1
not modelled
6.3
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rv2827c N-terminal domain-like99 c3m1eA_
not modelled
6.2
13
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator benm;PDBTitle: crystal structure of benm_dbd