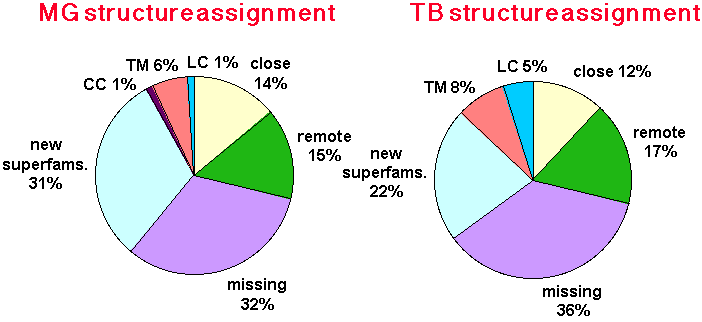
The pie-charts were constructed by first finding the fraction that can me annotated by close homologues and then by remote homologues only and finally by finding Coiled-coil, transmembrane and low complexity regions (only a few homologues of known structure overlap with these regions, this is because the protein structure database contains only a few transmembrane and coiled-coil proteins). Note that the fraction of coiled-coils in less than 1 percent in TB.We then calculated the expected fraction of missing assignments (undetected homologies) by multiplication of the 'remote' fraction with a factor of 2.1 (which is the ration of detected to undetected remote homologies as calculated in our benchmark). We believe the remaining part of the genome in new structural superfamilies.
Although both pie-charts look similar for both genomes the superfamily composition is rather different (see a table of the identified superfamilies for TB or MG).

Legend: CC (coiled-coils), TM (Transmembrane helices), LC (low complexety regions), close (machtes by close homologues), remote (matches by remote homologues only), missing (estimated undetected remote homologies) new superfams (fraction in potentially new structural superfamilies)
get structural annotations ...
Copyright © 1999-2002 Cancer Research UK
All Rights Reserved, disclaimer
Comments to author: a.mueller@cancer.org.uk
Generated: Thu Jun 27, 2002