| 1 | c3f9kV_ |
|
 |
99.8 |
18 |
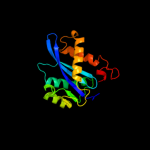
PDB header:viral protein, recombination
Chain: V: PDB Molecule:integrase;
PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
|
| 2 | d1c0ma2 |
|
 |
99.8 |
15 |
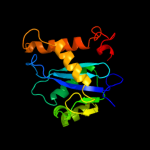
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 3 | c1c0mA_ |
|
 |
99.8 |
15 |
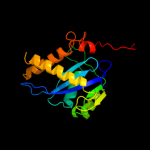
PDB header:transferase
Chain: A: PDB Molecule:protein (integrase);
PDBTitle: crystal structure of rsv two-domain integrase
|
| 4 | d1bcoa2 |
|
 |
99.8 |
17 |
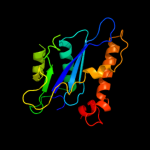
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:mu transposase, core domain |
| 5 | d1asua_ |
|
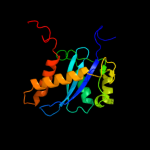 |
99.8 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 6 | d1exqa_ |
|
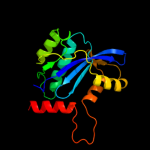 |
99.8 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 7 | c3nf9A_ |
|
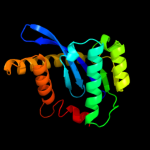 |
99.7 |
17 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:integrase;
PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
|
| 8 | c1ex4A_ |
|
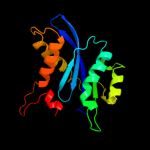 |
99.7 |
18 |
PDB header:viral protein
Chain: A: PDB Molecule:integrase;
PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
|
| 9 | c1bcoA_ |
|
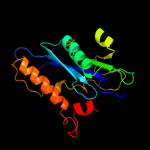 |
99.7 |
16 |
PDB header:transposase
Chain: A: PDB Molecule:bacteriophage mu transposase;
PDBTitle: bacteriophage mu transposase core domain
|
| 10 | c3kksB_ |
|
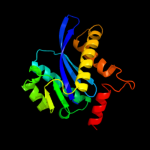 |
99.7 |
15 |
PDB header:dna binding protein
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
|
| 11 | d1c6va_ |
|
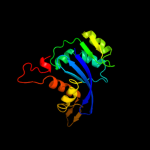 |
99.7 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 12 | d1cxqa_ |
|
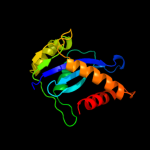 |
99.7 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 13 | c1k6yB_ |
|
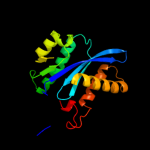 |
99.7 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
|
| 14 | d1hyva_ |
|
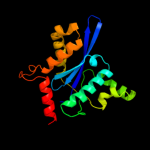 |
99.6 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 15 | c3hpgC_ |
|
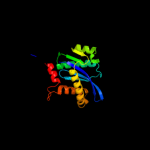 |
99.6 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:integrase;
PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
|
| 16 | c3l2tB_ |
|
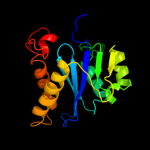 |
99.6 |
12 |
PDB header:recombination/dna
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
|
| 17 | c3dlrA_ |
|
 |
99.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:integrase;
PDBTitle: crystal structure of the catalytic core domain from pfv2 integrase
|
| 18 | c3hosA_ |
|
 |
98.7 |
17 |
PDB header:transferase, dna binding protein/dna
Chain: A: PDB Molecule:transposable element mariner, complete cds;
PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
|
| 19 | c2f7tA_ |
|
 |
97.4 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:mos1 transposase;
PDBTitle: crystal structure of the catalytic domain of mos1 mariner2 transposase
|
| 20 | c3f2kB_ |
|
 |
95.6 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:histone-lysine n-methyltransferase setmar;
PDBTitle: structure of the transposase domain of human histone-lysine2 n-methyltransferase setmar
|
| 21 | c3mwmA_ |
|
not modelled |
44.9 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:putative metal uptake regulation protein;
PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
|
| 22 | d1mzba_ |
|
not modelled |
43.2 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:FUR-like |
| 23 | c2fe3B_ |
|
not modelled |
41.5 |
30 |
PDB header:dna binding protein
Chain: B: PDB Molecule:peroxide operon regulator;
PDBTitle: the crystal structure of bacillus subtilis perr-zn reveals a novel2 zn(cys)4 structural redox switch
|
| 24 | c2o03A_ |
|
not modelled |
38.0 |
27 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable zinc uptake regulation protein furb;
PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
|
| 25 | c2fu4B_ |
|
not modelled |
36.5 |
29 |
PDB header:dna binding protein
Chain: B: PDB Molecule:ferric uptake regulation protein;
PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
|
| 26 | c2w57A_ |
|
not modelled |
36.3 |
33 |
PDB header:metal transport
Chain: A: PDB Molecule:ferric uptake regulation protein;
PDBTitle: crystal structure of the vibrio cholerae ferric uptake2 regulator (fur) reveals structural rearrangement of the3 dna-binding domains
|
| 27 | c2xigA_ |
|
not modelled |
34.2 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:ferric uptake regulation protein;
PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
|
| 28 | c3e7lD_ |
|
not modelled |
34.0 |
17 |
PDB header:transcription regulator
Chain: D: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
|
| 29 | c3eyyA_ |
|
not modelled |
33.3 |
31 |
PDB header:transport
Chain: A: PDB Molecule:putative iron uptake regulatory protein;
PDBTitle: structural basis for the specialization of nur, a nickel-2 specific fur homologue, in metal sensing and dna3 recognition
|
| 30 | c2kkmA_ |
|
not modelled |
28.5 |
15 |
PDB header:translation
Chain: A: PDB Molecule:translation machinery-associated protein 16;
PDBTitle: solution nmr structure of yeast protein yor252w [residues2 38-178]: northeast structural genomics consortium target3 yt654
|
| 31 | d2aq0a1 |
|
not modelled |
21.4 |
35 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 32 | d1f1ea_ |
|
not modelled |
19.4 |
20 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Archaeal histone |
| 33 | c1htrP_ |
|
not modelled |
12.4 |
26 |
PDB header:aspartyl protease
Chain: P: PDB Molecule:progastricsin (pro segment);
PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
|
| 34 | c1u78A_ |
|
not modelled |
10.8 |
7 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:transposable element tc3 transposase;
PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
|
| 35 | c1yshE_ |
|
not modelled |
10.7 |
19 |
PDB header:structural protein/rna
Chain: E: PDB Molecule:40s ribosomal protein s13;
PDBTitle: localization and dynamic behavior of ribosomal protein l30e
|
| 36 | d1gdta1 |
|
not modelled |
8.6 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 37 | c1s1hO_ |
|
not modelled |
8.4 |
31 |
PDB header:ribosome
Chain: O: PDB Molecule:40s ribosomal protein s13;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 38 | c3iwfA_ |
|
not modelled |
7.6 |
7 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator rpir family;
PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
|
| 39 | c3u5gK_ |
|
not modelled |
7.6 |
14 |
PDB header:ribosome
Chain: K: PDB Molecule:40s ribosomal protein s10-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
|
| 40 | c3oioA_ |
|
not modelled |
7.4 |
47 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (arac-type dna-binding domain-
PDBTitle: crystal structure of transcriptional regulator (arac-type dna-binding2 domain-containing proteins) from chromobacterium violaceum
|
| 41 | d1pdnc_ |
|
not modelled |
6.5 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Paired domain |
| 42 | d2gtad1 |
|
not modelled |
6.1 |
4 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |

